Research Article - Journal of Bacteriology and Infectious Diseases (2022) Volume 6, Issue 2
Isolation, identification and antibiotic susceptibility testing of salmonella from sheep and goats slaughtered at Haramaya Municipal Abattoir, Eastern Ethiopia.
Hordofa Noto, Feyera Gemeda*
Department of Veterinary Medicine, College of Agriculture and Veterinary Medicine, Jimma University, Jimma, Ethiopia
- *Corresponding Author:
- Feyera Gemeda
Department of Agriculture and Veterinary Medicine
Jimma University, Jimma, Ethiopia
E-mail: feyera.gemeda@ju.edu.et
Received: 03-Feb-2022, Manuscript No. AABID-22-53268; Editor assigned: 07-Feb-2022, PreQC No. AABID-21-53268(PQ); Reviewed: 21-Feb-2022, QC No. AABID-22-53268; Revised: 11-Mar-2022, Manuscript No. AABID-22-53268(R); Published: 18-Mar-2022, DOI:10.35841/aabid-6.2.108
Citation: Noto H, Gemeda F. Isolation, identification and antibiotic susceptibility testing of Salmonella from sheep and goats slaughtered at Haramaya Municipal Abattoir, Eastern Ethiopia. J Bacteriol Infec Dis. 2022;6(2):108
Abstract
A cross-sectional study was conducted from April to August 2021 at Haramaya municipal abattoir, eastern of Ethiopia with the aims to isolate, identify and to determine antimicrobial susceptibility of Salmonella. Isolation and identification was performed by conventional methods for detection and identification of Salmonella according to (ISO-6579, 2002). The antimicrobial susceptibility testing was performed by disc diffusion method. A total of 228 samples of meat, meat swab, cecum feces and skin swab was collected from sheep and goats, and examined for the presence Salmonella. Out of total samples, 34 (14.91%) was found positive for Salmonella and there was statistically significant variation between positive Salmonella and sample sources (p=0.000). All of 34 isolated Salmonella were exhibited 100% multi-drug resistance and highest percentages of resistance were observed for amoxicillin (100%), chloramphenicol (100%), ampicillin (94.1%) and tetracycline (70.6%). However, all isolates were susceptible to gentamicin and 97.1% were sensitive to kanamycin. The highest level of resistance of Salmonella against most commonly used antimicrobials detected by this study may pose challenges to veterinary and human health sectors. Therefore, it is advisable to work on improving a good hygiene in abattoir to minimize incidence of infection and it will be better if principle of antimicrobial stewardship applied and unregulated use of antibiotics avoided both in humans and animals.
Keywords
Antimicrobials, Disc diffusion, Haramaya, Salmonella, Susceptibility.
Introduction
Ethiopia is home for a large and diverse livestock resources and favourable production environments. The vast majority of the rural population’s livelihood is partly based on livestock production. The country had 59.5 million heads of cattle, 30.70 million heads of sheep, 30.20 million heads of goats, 56.53 millions of poultry and 1.21 million heads of a camel. It is central to the Ethiopian economy contributing about 45% to the agricultural GDP, supporting the livelihoods of 70 % of the population, 18.7% to the national GDP and 16-19% to the total foreign exchange earning of the country [1].
Meat is among the most valuable livestock products and for many people serve as their first-choice source of animal protein which provides all the essential amino acids and various micronutrients in proper proportion to the human beings. Meat composition makes it an ideal medium for the growth of a good number of microorganisms due to richness in nutrients. It is prone to contamination at various stages from primary production to when it is ready for consumption [2].
The genus Salmonella was named after Daniel E. Salmon who first reported the isolation of Salmonella from a pig in 1885 and named the organism Bacterium choleraesuis currently known as Salmonella enterica serovar Choleraesuis. Salmonellosis is an infectious disease of humans and animals caused by organisms of the genus Salmonella. Salmonellosis is one of the main foodborne zoonotic and animal husbandry problem throughout the world [3]. The bacteria cause foodborne poisoning in humans, mainly through animal products that include poultry, cattle, and pig products. Salmonella infections of food animals play an important role in public health and particularly in food safety, as food products of animal origin are considered to be the major source of human Salmonella infections [4].
The burden of foodborne illnesses is tremendous, affecting 10% of global population with 33 million deaths annually. Numerous factors contribute to diarrheal diseases, and Salmonella enterica causes foodborne illnesses with significant public health impact. Most Salmonella enterica serovars cause gastroenteritis that is a self-limiting infection that does not require antibiotic treatment [5].
Antimicrobial resistance is a global public health, animal health and welfare concern. Its development and spread is influenced by both human and animal antibiotic use. Veterinary antibiotics are medicines used to cure animals of bacterial infection [6]. Antibiotics used against bacteria are the most commonly recognized form of antimicrobials. Resistance is the ability of bacteria against the antagonizing effect of an antibacterial agent upon reproduction prevention or bactericidal. Antibiotics are used in food animal production to promote growth and to prevent (prophylactic), treat (therapeutic) and control (metaphylactic) infectious diseases [7]. The extensive use of antibiotics in the animal production systems for the purposes mentioned above has contributed to the development of drug-resistant bacteria. The close association of these bacteria has also been identified in the human food chain.
Antibiotic resistance in foodborne pathogens such as Salmonella is a major concern for public health safety. Antibiotic resistant Salmonella is a major global health concern owing to the increase in resistance to conventional antibiotics and the rise in multidrug resistance in recent years [8]. Infections that are difficult to treat or untreatable increase with AR, particularly as resistance to multiple antimicrobials increases. Bacteria, not humans or animals, become antibiotic-resistant. These bacteria may infect humans and animals, and the infections they cause are harder to treat than those caused by non-resistant bacteria [9].
Resistant bacteria are transferred from food animals to man via the food chain. Spread of resistance can involve the movement of resistant pathogenic bacteria themselves from one ecological niche to another (e.g., between animal and humans) or by indirect means (e.g., via the food chain and water supply) [10]. Pathogens and antibiotic resistance can arise from multiple sources, for example, the endemic pathogens circulating in the farm or pathogens introduced through the feed, water, workers, and equipment. Slaughterhouse is one of the most important risk factors that can act as a mixing vessel for any kinds and numbers of pathogens including Salmonella present in animals collected from different unrelated farms. Under favorable circumstances, such pathogens can be disseminated via the meat to the consumers [11, 12].
Farm and slaughterhouse workers, including veterinarians, have a high risk of being colonized or infected with resistant bacteria via direct contact with infected or colonized food animals and derived products [12]. Salmonella enterica have assumed epidemiological importance due the large number of outbreaks and infections caused by contaminated water and food consumption [13].
Antimicrobial Resistance (AMR) both in human and veterinary medicine has reached alarming levels in most parts of the world and has now been recognized as a significant emerging threat to global public health and food security [14]. Currently, the prevalence of antimicrobial resistant pathogens has increased at a speed inversely proportional to the approval of new drugs. Antimicrobial use has been identified as one of the driving factors behind the increase in AR, and use of antimicrobials in animals has been the subject of much debate [15].
In Ethiopia, large populations of livestock are kept and similarly large populations of humans are in contact with animals and/or with animal products [16]. Abattoir is among the places where animals are collected together from different farms or areas and there might be possibilities of contamination of meat and abattoir environment from sub-clinically ill animals when appropriate examinations are not employed on animals to be slaughtered. As a result, zoonotic pathogens can be transmitted to humans via ingestion or contact with contaminated meat and can cause public health impacts [17] In Haramaya woreda, like other parts of Ethiopia, different antibiotics are used to treat Salmonella in animals and it may be resistant to these antibiotics. And there was scarce information on prevalence and antibiotic resistant Salmonella from animals slaughtered at Haramaya municipal abattoir, eastern of Ethiopia. Therefore, the objectives of this study were
•To assess the prevalence of Salmonella from sheep and goats slaughtered in Haramaya municipal abattoir
•To determine the antibiotic susceptibility of Salmonella isolates from slaughtered sheep and goats in Haramaya municipal abattoir
Literature Review
General overview of Salmonella
The genus Salmonella contains more than 2400 serotypes based on their somatic (O), flagellar (H) and occasionally capsular antigens. Salmonella are rod-shaped, Gram-negative, non-sporulating organisms. All Salmonellae except S. enterica serotype Pullorum and S. enterica serotype Gallinarum are motile. Motility is mediated by peritrichous flagella [18]. As other Enterobacteriaceae they are oxidase negative but catalase positive. They are non-lactose fermenter that is why the colonies of Salmonella species have pale-straw colour on MacConkey agar. Most of Salmonella species are H2S, indole and citrate positives. Salmonellosis is considered as one of the most important life threating bacterial zoonotic disease of human as well as animals. The majority of Salmonellae of veterinary importance belong to S. enterica subspecies enterica. The subspecies are further qualified by the serotype to give a final designation such as S.enterica subspecies enterica serotype Typhimurium. In humans, Salmonella enterica Typhi (S. Typhi) and Salmonella enterica Paratyphi (S. Paratyphi) cause typhoid fever and paratyphoid fever, respectively, while salmonellosis is an overarching term which includes invasive infection with all serovars of Salmonella, as well as the normally gut restricted infections of food poisoning [19].
Humans appear to be susceptible to all Salmonella serotypes, with typhoid fever caused by S. Typhi, a disease restricted to humans, and infections with other serotypes being food-borne zoonoses [20-23]. Whether a person develops disease following ingestion of Salmonellae from the environment depends upon the dose of organisms, the serotype of Salmonella, and the colonization resistance of the infected individual [24].
Pathogenesis: The bacteria adhere to enterocytes through fimbriae and colonize the small intestine. They then penetrate enterocytes, where further multiplication occurs before they cross the lamina propria. The virulence of Salmonella serotypes relates to their ability to invade and replicate in epithelial cells. Because Salmonellae are facultative intracellular organisms which survive in the phagolysosome of macrophages, they can dodge the bactericidal effects of antibody and complement. They continue to proliferate, both free and within macrophages [25]. Survival within macrophages is necessary for development of systemic disease. Many Salmonella infections do not progress further. Further multiplication ultimately leads to septicaemia, with localization of bacteria in many organs and tissues. This includes the spleen, liver, meninges, brain, and joints [26].
Following attachment to the surface of intestinal mucosal cells, the bacteria induce ruffling of cell membranes. This ruffling is part of the mechanism whereby the organisms are taken up into non-phagocytic cells and is now known to be one of the functions encoded by genes on SPI-1 [27, 28]. This pathogenicity island is found in all serotypes of S. enterica analysed to date and one of its major effectors is a Type III Secretion System (TTSS). The TTSS is a complex of proteins which forms a needle like structure for the transfer of virulence factors from the bacterium into the host cell. Other products transferred by the TTSS activate secretory pathways and alter ion balance within the cell. In addition, effector proteins result in neutrophil recruitment, and the resulting inflammation, together with the disturbance of fluid and ion balance causes diarrhea.
Transmission: Salmonellae are spread by direct or indirect means. Infected animals are the source of organisms which they excrete and infect other animals directly, or indirectly by contamination of the environment, mainly feed and water supplies [29]. It is primarily transmitted by the fecal-oral route, often through ingestion of contaminated food and water. Infection is acquired by ingestion of material contaminated with infected faeces from either clinically ill animals, or carrier animals. The carrier state is particularly important in the maintenance and transmission of the disease. Human salmonellosis is generally foodborne and is contracted through consumption of contaminated food of animal origin such as meat, milk, poultry and eggs.
Clinical features: The disease can be described as three syndromes: Septicaemia, acute enteritis, and chronic enteritis [30]. Septicaemia is the characteristic form in newborn foals and calves and guinea-pigs. Affected animals show profound depression, dullness, prostration, high fever, and death within 24-48 hours. Acute enteritis is the common form in adult animals of all species. There is a high fever with severe fluid diarrhoea, sometimes dysentery. Other signs include anorexia, and faeces having a putrid smell, containing mucus, and sometimes blood and fibrinous casts. Pregnant animals usually abort. In all species, severe dehydration and toxaemia occur; the animal becomes recumbent and dies in 2-5 days [31].
Diagnosis, isolation and identification of Salmonella: Diagnosis is based on the identification of the Salmonella either from faeces or from tissues collected aseptically at necropsy, environmental samples or rectal swabs, feedstuffs and food products. In cases of intestinal infection, fecal samples are collected. Fresh fecal samples are placed onto nutrient, for example, blood agar, and one or more selective media, including Xylose Lysine Desoxycholate Agar (XLD), MacConkey agar and brilliant green agar [32-35]. In systemic or septicaemic disease, a blood sample is collected for standard blood culture. Spleen and bone marrow are cultured for the Salmonellae when postmortem diagnosis of systemic salmonellosis is required. Organism may be identified using a diversity of techniques that may include pre-enrichment to resuscitate sublethally damaged Salmonellae, enrichment media that comprise inhibitory substances to inhibit competing organisms, and selective agars to differentiate Salmonellae from other enterobacteria. Various biochemicals, serological and molecular tests can be used to the pure culture to allow for a reliable verification of an isolated strain [36]. Molecular techniques are now frequently used for the detection of Salmonellae in clinical and environmental samples, a major advantage being the speed with which a result can be obtained. PCR and real-time PCR-based tests can be applied directly to samples. Serological tests such as ELISA and agglutination techniques are of greatest value when used on a herd or flock basis.
Treatment: Antibiotic therapy should be based on results of susceptibility testing because R-plasmids coding for multiple resistance are comparatively common in Salmonellae [37]. Nursing care and appropriate antimicrobial therapy is the principal treatment for the enteric and systemic form of salmonellosis. Since Salmonellae survive in the phagocytic cell, the antimicrobial drug should be one that penetrates the cell. Examples of those that distribute in this manner include ampicillin, amoxicillin, gentamicin, trimethoprim–sulfonamides, and chloramphenicol/florfenicol. Fluid and electrolyte replacement therapy is required to counteract dehydration and shock.
Antibiotic resistance
Antibiotics are chemical agents that prevent bacterial growth by stopping the bacterial cells from dividing or by killing them [38-40]. The term Antibiotic Resistance (AR) is used to refer to the ability of bacteria to withstand the effect of one or more antibiotic agents at clinically attainable concentrations, usually resulting in therapeutic failure. Antibiotic resistance occurs when a bacterium that is normally susceptible to an antibiotic becomes able to grow in the presence of antibiotic levels that would normally suppress growth or kill susceptible organisms. Clinical resistance occurs when the bacterium can continue to divide in the presence of the antibiotic concentrations that normally occur during treatment (therapeutic doses) and the antibiotic is no longer effective for treatment [41].
Detecting methods of antibiotic resistance
Antibiotic susceptibility testing (AST) methods are in vitro procedures used to detect antibiotic resistance in individual bacterial isolates. AST determine or predict which antibiotic will be most successful in treating a bacterial infection in vivo. Those laboratory-based detection methods can determine resistance or susceptibility of an isolate against any therapeutic candidates. Those methods can also be used for monitoring the emergence and spread of resistant microorganisms in the population [42-45].
Susceptibility testing can involve phenotypic testing (i.e., determining the growth response of the organism of concern when exposed to the antibiotic) and genotypic testing (using polymerase chain reaction [PCR] to detect antimicrobial resistance genes of interest or whole genome and plasmid sequencing to detect the presence of antimicrobial resistance genes). Some examples of antibiotic sensitivity testing methods are dilution method (broth and agar dilution method) and disk-diffusion method. Among the available tests, agar disk-diffusion and the broth micro dilution methods are the two most commonly used methods in veterinary laboratories [46,47].
It is essential to follow standardized procedures to ensure results are repeatable and that results from one laboratory are comparable to those from another for the compilation of common data. Guidelines and recommendations for these are continuously updated by certain organizations worldwide, those which specify antimicrobial testing methods and interpretative criteria for veterinary pathogens are: the Clinical Laboratory Standards Institute (CLSI) in the USA, OIE in EU and Calibrated Dichotomous Sensitivity (CDS-AST) in Australia [48-51].
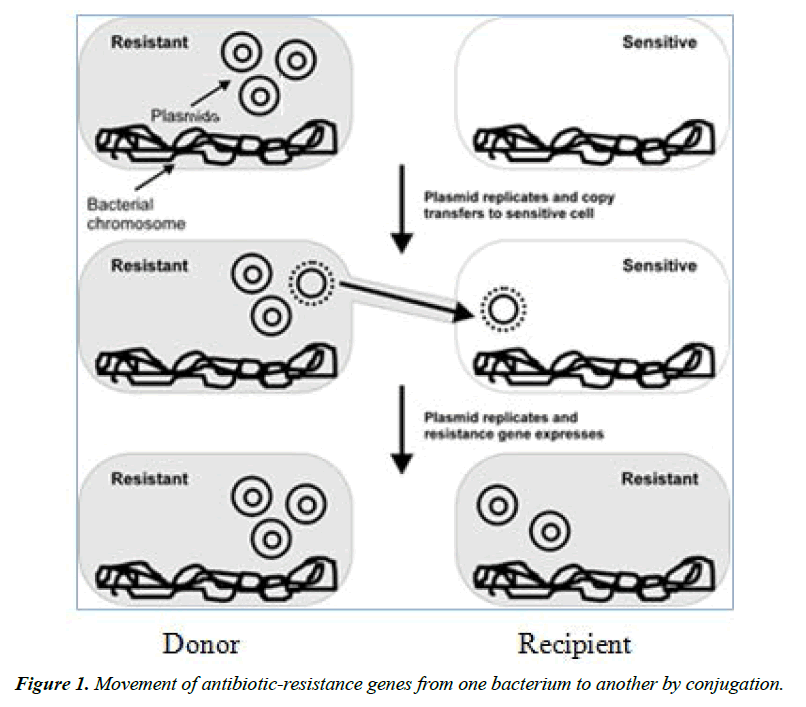
Disk-diffusion method: The application of commercially available drug-impregnated filter paper disks to the surface of an agar plate that has been inoculated to confluence with the organism of interest is called disk diffusion. Disc diffusion methods use filter paper discs impregnated with specific concentrations of antibiotics. The drug diffuses radially through the agar, the concentration of the drug decreasing logarithmically as the distance from the disk increases and results in a circular zone of growth inhibition around the disk, the diameter of which is inversely proportional to the MIC (Figure 1).
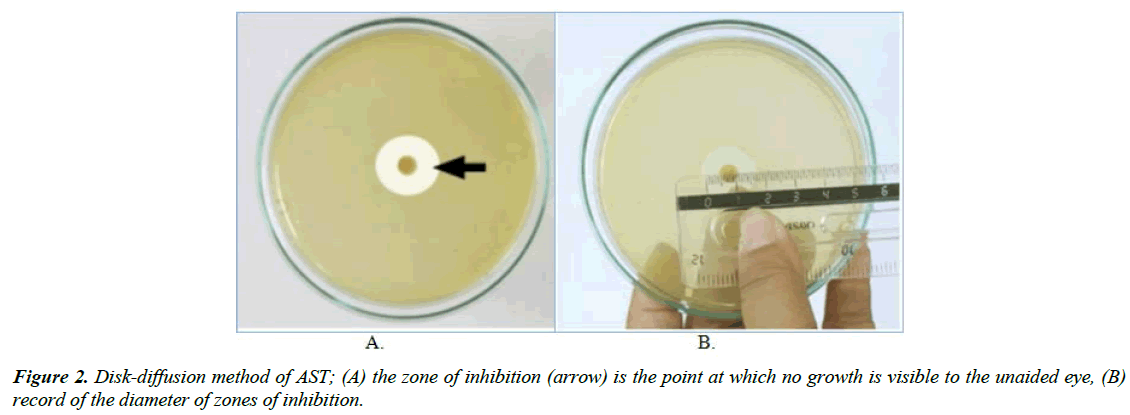
The diameter of the zone shows the susceptibility of the isolate and the diffusion rate of the drug through the agar medium. The diameter of the zone of growth inhibition is measured with calipers or a ruler and recorded in millimetres (Figure 2). Zones of growth inhibition around each of the antibiotic disks are measured to the nearest millimetre. The zone diameters are interpreted on the basis of guidelines published by CLSI, and the organisms are reported as susceptible, intermediate, or resistant [52].
Dilution method: The aim of the broth and agar dilution methods is to determine the lowest concentration of the antimicrobial that inhibits the visible growth of the bacterium being tested in either broth or on agar. Agar dilution and broth dilution are the most commonly used techniques to determine the minimal concentration of antibiotics that kill (bactericidal activity, MBC) or inhibit the growth (bacteriostatic activity, MIC) of bacteria. For both broth dilution methods, the lowest concentration at which the isolate is completely inhibited is recorded as the minimal inhibitory concentration or MIC. In clinical practice, this in vitro parameter is used to classify the tested microorganism as clinically susceptible, intermediate or resistant to the tested drug. Dilution methods are considered as reference methods for in vitro susceptibility testing and are also used to evaluate the performance of other methods of susceptibility testing [53,54].
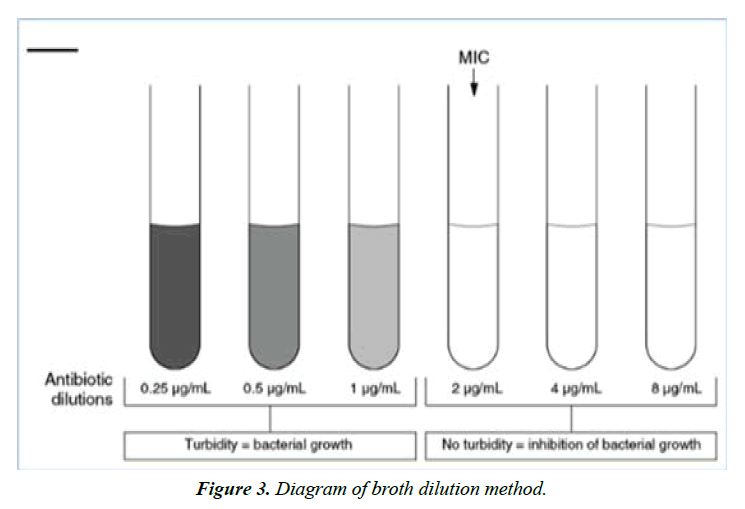
Broth dilution: The broth dilution technique of antibiotic susceptibility testing is also known as the Minimal Inhibitory Concentration (MIC) technique. Broth dilution uses liquid growth medium containing geometrically increasing concentrations (typically a twofold dilution series) of the antimicrobial agent, which is inoculated with a defined number of bacterial cells. It involves subjecting the isolate to a series of concentrations of antimicrobial agents in a broth environment. The concentration of the antibiotic in each tube is double that in the previous tube (Figure 3). The final volume of the test defines whether the method is termed macrodilution, when using a total volume of 2 ml, or microdilution, if performed in microtiter plates using ≤ 500 µl per well. Tubes are incubated under optimum conditions for the test microorganism from 16 to 24 hours. After incubation, the presence of turbidity or sediment indicates growth of the organism. Antimicrobial effect could be determined by spectrophotometry or by plating counting [55-58].
Agar dilution: Agar dilution involves the incorporation of varying concentrations of antimicrobial agent into an agar medium, usually using serial twofold dilutions, followed by the application of a defined bacterial inoculum to the agar surface of the plate. For agar dilution, solutions with defined numbers of bacterial cells are spotted directly onto the nutrient agar plates that have incorporated different antibiotic concentrations. After incubation, the presence of bacterial colonies on the plates indicates growth of the organism. The advantages of agar dilution testing include the reproducible results and satisfactory growth of most non-fastidious organisms. Agar dilution testing generally is not performed in routine clinical laboratories but can be ideal for regional reference laboratories or research laboratories that must test large numbers of isolates [42].
Prevention and control of antibiotic resistance
Antimicrobial resistance is a complex issue and requires human, animal and environmental health experts to work together to mitigate the continued development and spread of resistance. It is important to recognize that we cannot eliminate the emergence of resistance due to the rapid replication of bacteria, their ability to share resistance genes with other bacteria or to acquire them from their environment and any use of antimicrobials will continue to select for resistance. Therefore, our efforts must be focused on assuring that we are using antimicrobials as judiciously as possible and only in situations where the health or welfare of the patient would be compromised by a failure to treat.
Materials and Methods
Study area
The study area, Haramaya town, is situated in Haramaya woreda, East Hararghe zone, Oromia regional state, Ethiopia (Figure 4). It is located 21 km Northwest of Harar town and 505 km East of Addis-Ababa, capital city of Ethiopia. The altitude of this woreda ranges from 1400 to 2340 meters above sea level. It is characterized by “Woina-Dega” agro-climatic zone that receives mean annual rainfall of 775.9 mm. The monthly rainfall in the site is more than 100 mm from April to September, except June 48.4 mm. The wettest month is August, 151.9 mm. The daily temperature in the site ranges from 10 ºC to 25 ºC. The livelihood in the area is based on agriculture. The livestock population of Haramaya district is estimated at 71,205 heads of cattle, 15,294 sheep, 28,990 goats, 11755 donkeys, and 250 camels.
Study samples
The study samples were meat sample, meat swab, ceacal feces and skin swab of sheep and goats slaughtered at Haramaya municipal abattoir.
Study design
A cross-sectional study was conducted from April 2021 to August 2021 to determine the prevalence of Salmonella and its sensitivity to antibiotics on samples collected from sheep and goats slaughtered at Haramaya municipal abattoir.
Sample size determination: For isolation and identification of Salmonella, the sample size was determined and expected prevalence in sheep and goats samples with 5% desired absolute precision and 95% confidence interval using the formula recommended.
��= ��2×Pexp (1-Pexp)/��2, where
�� is required sample size, �� is 1.96, Pexp is expected prevalence, and �� is desired absolute precision of 0.05. Accordingly, the sample size was 196 but to increase the precision it was inflated by 16% and 228 samples was subjected to bacteriological examinations.
Sampling method, sample collection and transportation
The samples were collected randomly from slaughtered sheep and goats with proper labeling by sample type, sources and animal type (i.e., sheep or goat). All samples except skin swab (which is collected before slaughtering) were collected immediately after slaughter. The samples were collected from both sheep and goats slaughtered at Haramaya municipal abattoir during this study. Buffered peptone water, a transporting media, was used to transport the samples within required time to a laboratory. Each sample was placed within sample collection container depending on its type (e.g., feces, skin swab, meat sample, meat swab) (Table 1 and Table 2).
| Salmonella species | Hosts | Consequences of infection |
|---|---|---|
| Salmonella Typhimurium | Cattle, horses, sheep and goats, pigs, dogs and cats, poultry | Enterocolitis and septicaemia |
| Humans | Food poisoning | |
| Salmonella Dublin | Cattle, sheep and goats, horses, dogs, pigs | Abortion, Subclinical faecal excretors, Enterocolitis and septicaemia |
| Salmonella Choleraesuis | Pigs | Enterocolitis and septicaemia |
| Salmonella Pullorum, S. gallinarum |
Chicks, Adult birds | Pullorum disease, Fowl typhoid |
| Salmonella Anatum | Cattle, poultry, dogs and cats, horses | Septicemia, diarrhea, abortion |
| Salmonella Arizonae | Turkeys, sheep and goats | Arizona or paracolon infection |
| Salmonella Enteritidis | Poultry | Often subclinical in poultry |
| Humans | Food poisoning | |
| Salmonella Newport | Cattle, horses | Fever and diarrhea, weakness |
Table 1. Salmonella species of clinical importance and the consequences of infection.
| Antibiotics | Mode of action | Resistance mechanisms | Resistance genes |
|---|---|---|---|
| β-lactams- e.g., penicillins, cephalosporins |
Inhibits cell wall synthesis | β-Lactamases, Modification of porin (ompF), Efflux of β-lactam(ompC) | ompC, ompF, blaOXA-1, |
| Aminoglycosides-e.g., Neomycin, Kanamycin Gentamycin, Streptomycin |
Inhibits protein synthesis | Enzymatic modification and inactivation of aminoglycoside | aacC(3), aacC(3’), aadA, strA, strB |
| Phenicols e.g., Chloramphenicol, |
Inhibits protein synthesis | Efflux pumps(floR, cmlA),chloramphenicol acetyltransferase | floR, cmlA, cat1 |
| Tetracyclines | Inhibits protein synthesis | Efflux pumps, Modification of rRNA target, Inactivation of compound | tet(A),tet(B), tet(C), tet(D), tet(G), tet(H) |
| Sulfonamides | Inhibits metabolism | Dihydropteroate synthase | Sul1, sul2 sul3 |
Table 2. Mode of action and resistance mechanisms of antibiotics used to treat Salmonella.
For skin and meat swab sampling, approximately a 10×10 cm area was sampled. Skin swab samples were sampled from thigh, abdomen and thoracic area. For meat swab, abdomen (flank), thorax (lateral), and breast were sampling sites. Sterile cotton tipped swab that fitted with wooden shaft was first soaked in 10 ml of buffered peptone water and rubbed over the sampling area of meat and skin horizontally and then vertically many times. At the end of rubbing process, the wooden shaft of soaked cotton swab was broken off by pressing against inside wall and cotton swab was left in test tube containing buffered peptone water media. The meat samples from cervical (neck), abdomen (flank) and breast were collected in screw-cap jar containing buffered peptone water (BPW). Cecum feces was placed in test tube containing 10 ml of buffered peptone water and finally packed into ice box containing ice packs to transport samples to Veterinary microbiology laboratory of Haramaya University.
Isolation and identification of Salmonella
Isolation and identification of Salmonella was performed by conventional methods for detection and identification of Salmonella according to (ISO-6579, 2002) (Annex 1.1). The samples was pre-enriched in Buffered peptone water media, a non-selective pre-enrichment liquid media, and then incubated at 370 C for 24 hours. Then, 0.1 ml of pre-enriched samples were transferred into 10 ml of Rappaport-Vassiliadis soya (RVS) broth and incubated at 420 C for 24 hours. After incubation, a loop-full of selectively enriched culture from RVS broth was streaked onto the surface of Xylose lysine deoxycholate (XLD) agar media and incubated at 370 C for 24 hours.
Following incubation, presence of typical and suspected Salmonella colonies was examined on XLD plate. The presence of pink colonies with black centers, the typical Salmonella colonies on XLD plate, was subjected to biochemical tests for identification after subculture on nutrient agar (Annex 1.2). All colonies of presumptive Salmonella were sub cultured onto nutrient agar and incubated at 370 C for 24 hours and further identified by conventional biochemical tests such as Triple sugar iron (TSI) agar, Simmon’s citrate agar, Urease, methyl red (MR), Voges-Proscauer (VP) and indole test.
Antimicrobial susceptibility tests
The antimicrobial susceptibility testing of Salmonella isolates was performed by Kirby-Bauer disc diffusion method on Mueller-Hinton agar (Oxoid, England) according to the recommendations of. Biochemically confirmed Salmonella colonies that grown on nutrient agar were transferred into test tubes containing 0.8% saline solution until it have achieved 0.5 McFarland turbidity standards. Sterile cotton swab was dipped into the suspension, rotated several times and the bacteria swabbed uniformly over the surface of Mueller-Hinton agar plate (Oxoid, England). The plates were held at room temperature for 15-30 minutes to allow drying. Antibiotic discs with known concentration were dispensed by disc dispenser and the plates were incubated at 370 C for 24 hours. The isolates were tested for the susceptibility of the following antibiotic discs: Ampicillin (AMP) 10 µg, Amoxicillin-Clavulanic acid (AML) 25 µg, Gentamicin (GM) 10 µg, Kanamycin (K) 30 µg, Tetracycline (TE) 30 µg, Chloramphenicol (C) 30 µg, Nitrofurantoin (F) 300 µg and Erythromycin (E) 15 µg, discs were placed 23 mm apart and from plate edge. The plates were incubated at 370 C for 24 hours and interpretation of break points was recorded according to (Annex 1.3 and Annex 1.4).
Results
Prevalence of Salmonella
A total of 228 samples were collected from sheep and goats slaughtered in Haramaya municipal abattoir for detection and identification of Salmonella. A bacteriological examination by conventional culture and biochemical test methods was employed on 116 samples from sheep (meat sample, meat swab, skin swab and feces from cecum; each n=29) and 112 samples from goat (meat sample, meat swab, skin swab and faces from cecum; each n=28) (Annex 1.5 and Annex 1.6).
Out of total samples collected and processed, 34 (14.91%) was found positive for Salmonella and statistically significant variation between positive Salmonella and sample sources was observed (p=0.000). From a total of 116 samples collected from sheep, 16 were positive for Salmonella isolates. Of these, 3 (10.3%), 5 (17.2%), 2 (6.9%), 6 (20.7%) were found to be Salmonella positive from meat sample, meat swab, skin swab and feces, respectively. Out of 112 samples collected from goats, 18 were found to be positive for Salmonella, 7 (25%) from meat sample and 11 (39.3%) on feces from cecum of goats (Table 3).
| Species | Type of sample | No Examined | Number of Salmonella isolate | |
|---|---|---|---|---|
| Number positive | Total prevalence | |||
| Ovine | Meat sample | 29 | 3(10.3%) | 1.32% |
| Meat swab | 29 | 5(17.2%) | 2.19% | |
| Skin swab | 29 | 2(6.9%) | 0.88% | |
| Feces | 29 | 6(20.7%) | 2.63% | |
| Caprine | Meat sample | 28 | 7(25.0%) | 3.07% |
| Meat swab | 28 | - | - | |
| Skin swab | 28 | - | - | |
| Feces | 28 | 11(39.3%) | 4.82% | |
| Total | 228 | 34 | 14.91% | |
Table 3. Prevalence of Salmonella and type of sample from sheep and goats slaughtered in Haramaya municipal abattoir.
Antimicrobial susceptibility test
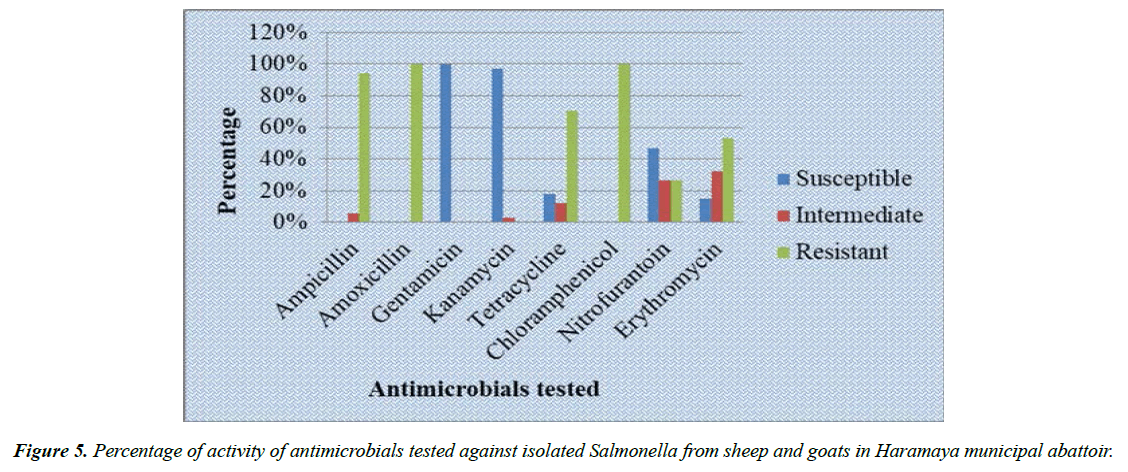
All of the 34 Salmonella isolates were subjected to eight antimicrobials to test its in vitro sensitivity. The highest resistance level (100% resistance) was observed for both amoxicillin and chloramphenicol. The next most frequent resistance was encountered to ampicillin, tetracycline and erythromycin with 32 (94.1%), 24 (70.6%) and 18 (52.9%) isolates being resistant, respectively (Figure 5). On the other hand, the isolates were 100%, 97.1% and 47% sensitive to gentamicin, kanamycin and nitrofurantoin respectively (Table 4 and Table 5). All of the 34 isolated Salmonella exhibited resistance to three or more antimicrobials, 100% multi-drug resistance was encountered (Table 6).
| Variables | Categories | No Examined | No Positive (%) | X2 (P- value) |
|---|---|---|---|---|
| Sample source | Meat sample | 57 | 10 (17.54%) | 17.836 (0.000) |
| Meat swab | 57 | 5 (8.77%) | ||
| Skin swab | 57 | 2 (3.5%) | ||
| Cecum feces | 57 | 17 (29.82%) | ||
| Species | Ovine | 116 | 16 (13.79%) | 0.233 (0.629) |
| Caprine | 112 | 18 (16.1%) | ||
| Sex | Male | 152 | 27 (17.76%) | 2.921 (0.087) |
| Female | 76 | 7 (9.21%) | ||
| Total | 228 | 34 (14.91%) |
Table 4. Prevalence of Salmonella in slaughtered sheep and goats on basis of sample source, species and sex categories.
| Antimicrobial drugs | Disc contents | Susceptible | Intermediate | Resistant |
|---|---|---|---|---|
| Isolate (%) | Isolate (%) | Isolate (%) | ||
| Ampicillin (AMP) | 10 µg | - | 2 (5.9%) | 32 (94.1%) |
| Amoxicillin (AML) | 25 µg | - | - | 34 (100%) |
| Gentamicin (GM) | 10 µg | 34 (100%) | - | - |
| Kanamycin (K) | 30 µg | 33 (97.1%) | 1 (2.9%) | - |
| Tetracycline (TE) | 30 µg | 6 (17.6%) | 4 (11.8%) | 24 (70.6%) |
| Chloramphenicol (C) | 30 µg | - | - | 34 (100%) |
| Nitrofurantoin (F) | 300 µg | 16 (47%) | 9 (26.5%) | 9 (26.5%) |
| Erythromycin (E) | 15 µg | 5 (14.7%) | 11 (32.4%) | 18 (52.9%) |
Table 5. Antimicrobial drugs and Salmonella isolates recovered from slaughtered sheep and goats that subjected to antimicrobials.
| Number of antimicrobial resistance | Antimicrobial resistance pattern (number of isolates) | Number of resistant isolates (%) |
|---|---|---|
| Zero/One/Two | None | 0 (0%) |
| Three | AML, AMP, C (3) AML, TE, C (1) AML, C, E (1) |
5 (14.7%) |
| Four | AML, TE, AMP, C (10) AML, AMP, F, C (1) AML, AMP, C, E (4) |
15 (44.12%) |
| Five | AML, AMP, F, C, E (1) AML, TE, AMP, F, C (1) AML, TE, AMP, C, E (6) |
8 (23.53%) |
| Six | AML, TE, AMP, F, C, E (6) | 6 (17.65%) |
Table 6. Multiple antimicrobial resistances of Salmonella isolated from slaughtered sheep and goats in Haramaya municipal abattoir.
Discussion
Since Salmonella is zoonotic pathogen and several antibiotic classes of the same family are used to treat salmonellosis in both veterinary and human medicine, surveillance on prevalence and antimicrobial resistance of Salmonella is essential to resolve and hinder the problem that will arise. In this study, the prevalence of Salmonella was 14.91%. The finding of this study is higher than that of studies conducted in Bishoftu (7.5%) Modjo (8.3%) and Addis Ababa (4.64%). The Bishoftu, Modjo and Addis Ababa abattoirs are export standard abattoirs and the current Haramaya municipal abattoir has poor sanitation and hygienic standard in comparison with export abattoirs. Therefore, the difference in prevalence of Salmonella could be due to the difference in sanitary and hygienic practices employed in the abattoirs, cross-contamination of carcasses with intestinal tract contents during slaughtering, water used and the hygiene of environment up on which the animals are slaughtered are important factors. And might be because of variation in animal feeding habits, types of feed provided, housing condition, sampling methods and culturing techniques.
However, the finding of this study was parallel with the studies conducted in Tigray region (16.4%), Wolaita Sodo (12.5%), Dire Dawa (17.7%) and Elfora and Luna export abattoirs (17.21%). In this study, out of 228 samples collected and processed, 34 (14.91%) was found to be positive for Salmonella. Higher prevalence was observed in goats (7.89%) than in sheep (7.02%), which was in contrary with finding of study conducted in which higher prevalence was in sheep (4.08%) than goats (0.85%). This difference may be due to stressing factors, animal management differences within and between study areas and also variation in study population of two species. However, this finding is supported in which prevalence was higher in goats (9.01%) than sheep (8.41%).
Out of 34 Salmonella isolates, 17 (7.45%) were found on feces, 10 (4.39%) on meat sample, 5 (2.19%) on meat swab and 2 (0.88%) on skin swab. Salmonella is carried in intestinal tract of animals and excreted in their feces especially during stresses such as transportation. This study reveals higher prevalence of Salmonella in feces (7.45%) in comparison to other sample sources and it is in line with finding in which Salmonella isolates are higher (5.73%) in cecum content. Hence in the abattoir, feces could be potential source of Salmonella for meat and environmental contamination and risk for abattoir workers. In the current study, Salmonella contamination was present on meat sample and swab at a level of 4.39% and 2.19%, respectively. This level of meat contamination is lower as compared to the 12.5% and 17.7% prevalence in study conducted, respectively. However, a report indicated 2.5% prevalence of Salmonella on carcass swab, which is in line with result of this study. This carcass contamination is public health issue for a country like Ethiopia, where there is a culture of eating raw and/or undercooked meat.
As compared to other enteric microbes, Salmonella most frequently present on animals body coat. In current study, the occurrence of Salmonella on skin of sheep in abattoir was 0.88%. The proportion of Salmonella on skin was lower compared to a study reported 8% at farm and 25% prevalence in slaughterhouse in Pakistan, 31% and 7.1% prevalence on hide in Ethiopia, respectively. This difference could be due to overcrowding of animal in lairage and hair coat contamination during transportation. Thus, presence of Salmonella on animal body coat (skin) can be a source of infection for individuals in contact with infected animals.
Antimicrobial Resistance (AMR) is an emerging problem and the most significant animal and public health challenge of this century globally. In the present study, all of the 34 Salmonella isolates were subjected to eight antimicrobials and each isolates were resistant to at least three antimicrobials, 100% Multiple Drug Resistance (MDR) was detected. High percentage of multi-drug resistant Salmonella isolates to commonly used antimicrobials observed in this study could pose challenge to both public health and longer use of effective antimicrobials. The finding of this study regarding 100% multiple drug resistance was higher than the study who reported 23.5%, 53.84%. However, the occurrence of 100% multiple drug resistant Salmonella isolates in this study is consistent with study in wich both of them reported 100% multi-drug resistant Salmonella in Ethiopia and Bangladesh, respectively.
Salmonella isolates from stool samples in Harar were resistant to commonly used antimicrobials including ampicillin, amoxicillin, tetracycline and chloramphenicol. The result of the current study also indicated resistance of Salmonella isolates to commonly used antimicrobials including amoxicillin, ampicillin, chloramphenicol, tetracycline, erythromycin and nitrofurantoin with resistance rate of 100%, 94.1%, 100%, 70.6%, 52.9% and 26.5%, respectively. All the isolated Salmonella, in present study, were exhibited 100% resistance to amoxicillin and chloramphenicol. Resistance to amoxicillin in 100% rate in this study was higher in which 42.9% resistance were reported, however, the current study is consistent with finding of study by reported 100% resistance. High resistance percentage (100%) to chloramphenicol in this study also higher than 69.23% reported and 51.8%, however, it is supported who reported 100% resistance of Salmonella isolate to chloramphenicol.
The resistance of isolated Salmonella to ampicillin in this study was 94.1%, which is in line with 100% resistance reported but disagree with reports in which 38.46% and 46.4% resistance reported respectively. This difference of resistance could be due to frequent and inappropriate utilization of antimicrobials both in humans and animals, which favours selection pressure that increase resistance genes in bacteria.
Gentamicin and kanamycin showed a good antimicrobial activity against isolated Salmonella. The sensitivity of all 34 isolates to gentamicin in this study is comparable with findings in Ethiopia in Kenya who reported 92.8%, 92.3% and 97% susceptibility respectively, but contradict with the study conducted in Jimma University specialized hospital and in Hossana in which both study reported 100% resistance against gentamicin. The high level (97.1%) of susceptibility of isolated Salmonella to kanamycin in this study is in agreement with study reported 100% susceptibility but higher than the findings, reported 41.7%, 25% and 17.9% susceptibility rate, respectively. The highest antimicrobial activity of gentamicin and kanamycin against isolated Salmonella in the present study maybe due to limited access and usage in veterinary and public health sectors compared to other antimicrobials in different parts of Ethiopia.
Conclusion
Salmonellosis is the main foodborne zoonotic and animal husbandry problem throughout the world. This study detected 14.91% overall prevalence of Salmonella in samples collected from slaughtered sheep and goats, which can significantly be a potential source of human salmonellosis. In this study, Salmonella was isolated from feces with 7.45% and 0.88% proportion in skin swab (body coat), which suggests Salmonella from feces and exterior of animal body coat can contaminate meat in abattoir during slaughtering process. The contamination of meat that observed with 4.39% in this study is also risk for consumers. All of the isolated Salmonella were exhibited 100% multi-drug resistance to antimicrobials that are used commonly in veterinary and human medicine, this pose risk to human and animal’s health. Generally, the level of antimicrobial resistance observed in this study gives us acues on how we have to use antibiotics. Thus, a judicious use of antibiotics in healthcare and animal health sector is essential to slow the emergence of resistance and extend the useful lifetime of effective antibiotics.
Therefore, depending up on the findings of this study, the following recommendations are forwarded in order to reduce the emergence and impacts of antimicrobial resistance:
•It is advisable to avoid cross contact of carcass with gastrointestinal contents in abattoir which will minimize the contamination of Salmonella from intestinal contents.
•The floor of abattoir up on which the animals are slaughtered should be cleaned, personal hygiene should be improved and potable water should also be used for washing purpose.
•It will be better if principle of antimicrobial stewardship applied and unregulated use of antibiotics avoided both in humans and animals.
•It is better if other studies concerning sources of Salmonella contamination in the abattoir is performed.
References
- Abebe M, Tafese B, Adane H. Antimicrobial resistance of Salmonella serovars isolated from food of bovine origin in selected Woredas of Tigray, Ethiopia. World J Med Sci 2014;11:342-47.
- Abe K, Jelalu K, Haile A, et al. Isolation, identification, and antibiotic susceptibility testing of Salmonella from slaughtered bovines and ovines in addis ababa abattoir enterprise, Ethiopia: A cross-sectional study. Int J Bacteriol 2016;2016:3714785.
- Abebe W, Alemu E, Solomon T, et al. Prevalence and antibiotic susceptibility patterns of Shigella and Salmonella among children aged below five years with diarrhoea attending nigist eleni mohammed memorial hospital, South Ethiopia. BMC Pediatr 2018;18:241.
- Abunna F, Jote H, Beyene T, et al. Isolation, identification and antimicrobial susceptibility profile of Salmonella isolates from abattoir and dairy farms in and around holeta town, oromia, Ethiopia. J Vet Med Res 2017;4:11113.
- Addis Z, Kebede N, Sisay Z, et al. Prevalence and antimicrobial resistance of Salmonella isolated from lactating cows and in contact humans in dairy farms of Addis Ababa: a cross sectional study. BMC Infect Dis 2011;11:222.
- Afolami OI, Onifade AK. Antibiotic resistant Salmonella spp: Mechanism of drug resistance, gene variations and clinical implications. Asian J Res Med Pharm Sci 2018;4:1-6.
- Aftab M, Rahman A, Qureshi MS, et al. Level of Salmonella in beef of slaughtered cattle at Peshawar. J Anim Plant Sci 2012;22:24-27.
- Akafete T, Haileleul N. Assessment of risk factors and prevalence of Salmonella in slaughtered small ruminants and environment in an export abattoir, Modjo, Ethiopia. American-Eurasian J Agric Environ Sci 2011;10:992-99.
- Alam SB, Mahmud M, Akter R, et al. Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens 2020;9(3):201.
- Alemu S, Zewde BM. Prevalence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from slaughtered cattle in Bahir Dar, Ethiopia. Trop Anim Health Prod 2012;44:595-600.
- Ayalu AR, Berhanu S, Jemal Y, et al. Antibiotic susceptibility patterns of Salmonella and Shigella isolates in Harar, Eastern Ethiopia. J Infec Dis Immun 2011;3(8):134-39.
- Belay DE. Smallholder livestock production and marketing systems in the Haramaya district, eastern Ethiopia. Basic Res J Agri Sci Rev 2013;2(6):122-29.
- Bersisa A, Tulu D, Negera C. Investigation of bacteriological quality of meat from abattoir and butcher shops in Bishoftu, Central Ethiopia. Int J Microb 2019;2019:8.
- Bhandare, SG, Sherikar AT, Paturkar AM, et al. A comparison of microbial contamination on sheep/goat carcasses in a modern Indian abattoir and traditional meat shops. Food Control 2007;18(7):854-58.
- Cesur S, Demiroz AP. Antibiotics and the mechanisms of resistance to antibiotics: review article. Med J Islamic World Acad Sci 2013;21(4):138-42.
- Dawit GY, Walelegn WY, Shafi A. Food Safety practice and associated factors among meat handlers in gondar town: a cross-sectional study. J Environ Public Health 2020;2020: 7421745.
- Destaw AA, Belege T, Aragaw E. Prevalence and antibiotic resistance pattern of Salmonella isolated from caecal contents of exotic chicken in debre zeit and modjo, Ethiopia. Hindawi Int J Microbiol 2020.
- Divek VT. Nair KV, Johny AK. Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control: Review. Foods 2018;7:167.
- El-Ghany WA. Salmonellosis: A food borne zoonotic and public health disease in Egypt. J Infect Dev Ctries 2020;2018:14:674-78.
- Feyisa K, Matios L, Tafesse K, et al. A cross sectional study on Salmonella in apparently healthy sheep and goats slaughtered at Elfora and Luna export abattoirs, Ethiopia. African J Microb Res 2017;11(13):530-36.
- Frye JG, Charlene JR. Genetic mechanisms of antimicrobial resistance identified in Salmonella enterica, Escherichia coli, and Enteroccocus spp. isolated from U.S. food animals. Front Microbiol 2013;4:135.
- Frye JG, Rebecca LL, Richard JM, et al. Related antimicrobial resistance genes detected in different bacterial species co-isolated from swine fecal samples. Foodborne Pathog Dis. 2011;8(6):663-79.
- Glenn LS, Rebecca LL, Joseph FF, et al. Analysis of antimicrobial resistance genes detected in multidrug-resistant Salmonella enterica serovar typhimurium isolated from food animals. Microb Drug Resist 2011;17(3):407-18.
- Harriet U, Nandita De. Mechanisms of Antibiotic resistance in Salmonella typhi: Review Article. Int J Curr Microb App Sci 2014;3(12):461-76.
- Hendry SA, Dennis J. Bacterial culture and antibiotic susceptibility testing. Compend Contin Educ Vet 2010;32(7):1-5.
- Jiang Z, Paudyal N, Xu Y, et al. Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in henan, china. Front Microbiol 2019;10(1513):1-9.
- Jones RN, Ballow CH, Biedenbach DJ, et al. Multi-laboratory assessment of the linezolid spectrum of activiti using the Kirby-Bauer disk diffusion method: Report of the Zyvox Antimicrobial Potency Study (ZAPS) in the United States. Diagn Microbiol Infect Dis 2001;40(1-2):59-66.
- Kagambega LT, Aulu L, Traore AS, et al. Prevalence and characterization of Salmonella enterica from the feaces of cattle, poultry, swine and hedgehogs in Burkina Faso and their comparison to human Salmonella isolates. BMC Microbiol 2013;13: 2-9.
- Kemal J, Sibhat B, Menkir S, et al. Prevalence, assessment, and antimicrobial resistance patterns of Salmonella from raw chicken eggs in Haramaya, Ethiopia. J Infect Dev Ctries 2016;10(11):1230-35.
- Kumar S, Singh BR. An overview of mechanisms and emergence of antimicrobials drug resistance. Adv Ani Vet Sci 2013;1(2S):7-14.
- Lambertini E, Ruzante JM, Chew R, et al. The public health impact of different microbiological criteria approaches for Salmonella in chicken parts. Microbial Risk Analysis 2019;12:44-59.
- Landers TF, Bevin C, Thomas E, et al. A Review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep 2012;127:4-22.
- Li R, Lai J, Wang Y, et al. Prevalence and characterization of Salmonella species isolated from pigs, ducks and chickens in Sichuan Province, China. Int J Food Microbiol 2013;163:14-18.
- Lidya K, Zerihun K, Bitsu K, et al. Prevalence and antimicrobial susceptibility profile of Salmonella serovars isolated from slaughtered cattle in addis Ababa, Ethiopia. BioMed Res Int 2018;2018:9794869.
- Mathew AG, Cissell R, Liamthong S. Antibiotic resistance in bacteria associated with food animals: a United States perspective of livestock production. Food Borne Path Dis 2007;4:115-33.
- McEwen SA, Fedorka-Cray PJ. Antimicrobial use and resistance in animals. Clin Infect Dis. 2002;34:3:S93-S106.
- McGeer A. Agricultural antibiotics resistance in human pathogens: villain or scapegoat?. CMAJ 1998;159(9):119-20.
- Mutai CW, Muigai AW, Waiyaki P, et al. Multi-drug resistant Salmonella enterica serovar Typhi isolates with reduced susceptibility to ciprofloxacin in Kenya. BMC Microbiol 2018;18:187.
- OIE. Laboratory methodologies for bacterial antimicrobial susceptibility testing. OIE Terresterial Manual 2019;8.
- Pui CF, Wong WC, Chai LC, et al. Salmonella: A foodborne pathogen. Int Food Res J 2011;18:465-73.
- Shimels TY. Review on Antibiotic Resistance: Resistance Mechanisms, Methods of Detection and Its Controlling Strategies. Biomed J Sci & Tech Res 2020;24(5):18651-18657.
- Shishaye H, Nagari A. Hydrogeochemical analysis and evaluation of the groundwater in the haramaya well field, eastern hararghe zone, Ethiopia. J Hydrogeology & Hydrologic Engineering 2016;5(4):1000146.
- Sibhat B, Molla ZB, Zerihun A, et al. Salmonella serovars and antimicrobial resistance profiles in beef cattle, slaughterhouse personnel and slaughterhouse environment in Ethiopia. Zoonoses Public Health, Vol 2011;58:102-09.
- Silva KC, Knobl T, Moreno AM. Antimicrobial resistance in veterinary medicine: mechanisms and bacterial agents with the greatest impact on human health. Brazilian J Vet Res Ani Sci 2013;50(3):171-83.
- Sime MG. Occurrence and Antimicrobial Susceptibility of Salmonella in Fecal and Carcass Swab Samples of Small Ruminants at Addis Ababa Livestock Market. J Vet Sci Technol 2021;12(2).
- Tadesse G, Tessema TS. A meta-analysis of the prevalence of Salmonella in food animals in Ethiopia. BMC Microbiol 2014;14:270.
- Threlfall E. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol Rev 2002;26:141-48.
- Tizazu Z, Subbaram K, Daniel Y, et al. Invasive Bacterial Pathogens and their Antibiotic Susceptibility Patterns in Jimma University Specialized Hospital, Jimma, Southwest Ethiopia. Ethiop J Health Sci 2011;21(1):1-8.
- Van Honert MS, Gouws PA, Hoffman LC. Importance and implications of antibiotic resistance development in livestock and wildlife farming in South Africa: A Review. S Afr J Anim Sci 2018;48(3):1.
- Van Duijkeren E, Anne-Kathrin S, Marilyn CR, et al. Mechanisms of bacterial resistance to antimicrobial agents. Am Soc Microbiol 2018.
- Van Hoek AH, Mevius D, Beatriz G, et al. Acquired Antibiotic Resistance Genes: An Overview. Front Microbiol 2011;2:203.
- Wales AD, Cook AJ, Davies RH. Producing Salmonella-free pigs: a review focusing on interventions at weaning. Vet Rec 2011;168(10):267-76.
- Wang X, Biswas S, Paudyal N, et al. Antibiotic Resistance in Salmonella Typhimurium Isolates Recovered From the Food Chain Through National Antimicrobial Resistance Monitoring System Between 1996 and 2016. Front microbiol 2019;10(985).
- Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc, 2008;3(2):163-75.
- Woldemariam E, Molla B, Alemayehu D, et al. Prevalence and distribution of Salmonella in apparently healthy slaughtered sheep and goats in Debre Zeit, Ethiopia. Small Ruminant Res 2005;58:19-24.
- Wondimu W, Abraham Y, Anjulo AA. Detection and identification of antimicrobial-resistant Salmonella in raw beef at Wolaita Sodo municipal abattoir, Southern Ethiopia. J Health Popul Nutr 2017;36:52.
- Yan SS, Pendrak ML, Abela-Ridder B, et al. An overview of Salmonella typing: public health perspectives. Clin Appl Immunol 2003;4:189-204.
- Zewdu E, Cornelius P. Antimicrobial resistance pattern of Salmonella serotypes isolated from food items and personnel in Addis Ababa, Ethiopia. Trop Anim Health Prod 2009;41:241-49.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref