Research Article - Biomedical Research (2016) Volume 27, Issue 4
Ethanol extract of Cimicifugae rhizoma exerted more potent antiinflammatory and tumor suppressor activities compared with methanol and water extracts
Su-Hyeon Jeong1, Huifang Guo2, Hye-Young Kim3* and Jun-Beom Park4*1Department of Rehabilitation Medicine of Korean Medicine, Chungju Hospital of Korean Medicine, College of Korean Medicine, Semyung University, Jecheon, 27429, Republic of Korea
2Department of Medical Biotechnology, College of Biomedical Science, Kangwon National University, Chuncheon-si, 24341, Republic of Korea
3Department of Dental Hygiene, College of Health Science, Kangwon National University, Samcheok-si, 25949, Republic of Korea
4Department of Periodontics, College of Medicine, The Catholic University of Korea, Seoul, 06591, Republic of Korea
- *Corresponding Author:
- Hye-Young Kim
Department of Dental Hygiene
College of Health Science
Kangwon National University
Republic of Korea
Jun-Beom Park
Department of Periodontics
College of Medicine
The Catholic University of Korea
Republic of Korea
Accepted date: March 24, 2016
Abstract
Cimicifugae rhizoma (C. rhizoma) called Shengma (Chinese), Seungma (Korean), and Shoma (Japanese) is primarily derived from Cimicifuga heracleifolia Komarov or Cimicifuga foetida Linnaeus. C. rhizoma is used for anti-inflammatory, analgesic, antipyretic remedy and alternative for hormone replacement therapy. The aim of this study was to determine the dose-dependent anti-inflammatory and tumor suppressor activities using methanol, ethanol and water extract of C. rhizoma. Methanol, ethanol and water extracts of Cimicifuga heracleifolia Komarov were obtained. Mouse leukaemic monocyte macrophage cell line (RAW 264.7) was loaded in the presence of C. rhizoma at final concentrations that ranged from 10 to 50 μg/mL. The production of Nitric Oxide (NO) in lipopolysaccharides-induced RAW 264.7 cells was quantified. Cycloxygenase-2 (COX-2) mRNA expression was evaluated using Lipopolysaccharides (LPS)-stimulated RAW 264.7 cells with semiquantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR). Lung carcinoma cell lines (A549) were treated with C. rhizoma at final concentrations that ranged from 200 to 800 μg/mL and expressions of caspase-3 and Superoxide Dismutase-2 (SOD-2) were tested with RT-PCR. No changes of cell viability were noted after treatment with methanol, ethanol, and water extracts of C. rhizoma. Stimulation with LPS for 24 h led to a robust increase in the NO production, but C. rhizoma significantly suppressed NO by the LPS-stimulated RAW 264.7 cells in all groups. Pretreatment with absolute ethanol extract of C. rhizoma suppressed the LPS-stimulated COX-2 expression. The results showed that C. rhizoma extract significantly showed anticancer effects in methanol and ethanol groups. The expression of caspase-3 and SOD-2 increased with the increase of exposure time with C. rhizoma extract. Within the limits of this study, C. rhizoma showed anti-inflammatory effect using RAW 264.7 cell line and tumor suppressor activities on A549 cells. Absolute ethanol extract showed the highest antiinflammatory and tumor suppressor activities on these experimental settings.
Keywords
Anti-inflammatory agents, Antineoplastic agents, Herbal medicine, Plant roots.
Introduction
The genus Cimicifuga, belonging to the family Ranunculaceae, is a group of perennials which are distributed widely in Asia, Europe and United States [1]. Cimicifugae rhizoma (C. rhizoma) called Shengma (Chinese), Seungma (Korean), and Shoma (Japanese) is primarily derived from Cimicifuga heracleifolia Komarov or Cimicifuga foetida Linnaeus [2]. C. rhizoma is a traditional herb medicine that is used to treat various diseases as an anti-inflammatory, analgesic, antipyretic remedy and alternative for hormone replacement therapy [1,3-6]. It has been clinically used for the treatment of headache, generalized fever, oral erosion, prolapse of rectum and sore [1].
The anticancer properties of Genus Cimicifuga have been revealed [7]. Ethyl acetate fraction from the aerial part of Cimicifuga foetida Linnaeus possessed the anti-tumor action on hepatoma, and the fraction inhibited the growth of the implanted mouse H22 tumor in a dose-dependent manner with the growth inhibitory rate of 63.32% at 200 mg/kg [8].
Extracts C. rhizoma using different solvent including methanol and water were tested for anti-inflammatory effects in the previous study [1]. The aim of this study was to determine the dose-dependent anti-inflammatory and tumor suppressor activities using methanol, ethanol and water extract of C. rhizoma. To our knowledge, this investigation is the first to elucidate the comparative effect of methanol, ethanol and water extract of C. rhizoma on mouse leukaemic monocyte macrophage cell line (RAW 264.7) and lung carcinoma cell lines (A549).
Materials and Methods
Chemicals
All chemicals used in this study were purchased from Sigma- Aldrich (St. Louis, MO, USA) unless specified.
Plant material and preparation of the extract
The dry roots of Cimicifuga heracleifolia Komarov were obtained from Chungju Hospital of Korean Medicine, College of Korean Medicine, Semyung University, Jecheon, Republic of Korea. The roots of Cimicifuga heracleifolia Komarov were chopped to small size of 0.5 cm long, dried in shade and powdered in mechanical grinder. The pulverized roots were extracted with absolute ethanol, 70% ethanol, absolute methanol, 70% methanol, water and boiling water at 60°C for 3 hours and finally the extraction was dried under vacuum rotary evaporator (CCA-1110; Eyela, Tokyo, Japan).
Thirty grams of dry roots in each group were used and 3.10, 6.45, 4.52, 6.35, 6.14 and 7.38 g was obtained for absolute ethanol, 70% ethanol, absolute methanol, 70% methanol, water and boiling water, respectively. The yield of absolute ethanol, 70% ethanol, absolute methanol, 70% methanol, water and boiling water groups were 10.33, 21.50, 15.07, 21.17, 20.47 and 24.60 % (w/w) respectively.
Anti-inflammatory assay
Cell line and cell culture: The RAW 264.7 cell line was purchased from the Korean Cell Line Bank (Seoul, Korea). RAW 264.7 cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 100 U/mL of penicillin, and 100 μg/mL of streptomycin. The cells were incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Cell cytotoxicity assay: The cytotoxicity of samples on RAW 264.7 cells was tested. Cells were seeded into 96-well plates at a density of 1 × 105 cells/well. After incubation for 18 h, cells were exposed to medium along with samples at different concentrations for 24 h. The supernatant was removed from each well and 10 μL of MTT solution (5 mg/mL in phosphatebuffered saline) and 90 μL of FBS-free medium were added to each well and incubated for 4 h incubation at 37°C. Then the supernatant was sucked out and 200 μL of DMSO was added into each well. The plate was vibrated slightly for 10 min and the amount of MTT formazan was quantified by measuring absorbance at 490 nm using an enzyme-linked immunosorbent assay (ELISA) plate reader (ELx800TM, Bio-Tek, Winooski, VT, USA). The optical density of formazan formed in the control cells was considered 100% viability. Cell viability was expressed as a percentage of the control culture value.
Quantification of NO production in Lipopolysaccharides (LPS)-induced RAW 264.7 cells: RAW 264.7 cells were plated in 96-well cell plates and incubated for 18 h. Then cells were stimulated with LPS (2 μg/mL) in the presence or absence of samples with various concentration for 24 h [9]. Aliquots of 100 μL of cell culture medium was mixed with 100 μL of Griess reagent [0.1% aqueous solution of naphthylethylenediamine dihydrochloride, 50 μL; 1% sulfanilamide (in 5% phosphoric acid), 50 μL]. The absorbance was determined at 550 nm using an ELISA plate reader (ELx800TM).
Reverse Transcription-Polymerase Chain Reaction (RTPCR) analysis: RAW 264.7 cells (1 × 106) were grown in 6- well plates for 18 h. Then cells were treated with various concentrations of sample for 30 min and LPS (2 μg/mL) was added. After incubation for 24 h, total RNA of the cells was isolated with a Trizol RNA isolation kit (Invitrogen, Carlsbad, CA, USA). The total RNA was reverse-transcribed to cDNA and used as the template for PCR amplification. The forward and reverse primers were as follows: 5'- CACTACATCCTGACCCACTT-3' and 5'- ATGCTCCTGCTTGAGTATGT-3' for cyclooxygenase-2 (COX-2), and 5'-CACTCACGGCAAATTCAACGGCA-3' and 5'-GACTCCACGACATACTCAGCAC-3' for GAPDH. The amplified PCR products were separated on 1% agarose gel, and the gel was stained with ethidium bromide. The gel was photographed with a Mini BIS Image Analysis System (DNR Bio-Imaging Systems Ltd., Jerusalem, Israel).
Tumor suppressor activity assay
Cell line and cell culture: The lung cancer A549 cell line was purchased from the Korean Cell Line Bank (Seoul, Korea). Cells were maintained in DMEM medium supplemented with 10% FBS (Fetal Bovine Serum), 100 U/mL of penicillin, and 100 μg/mL of streptomycin. The cells were incubated at 37°C in a humidified atmosphere of 95% air and 5% CO2.
Cytotoxicity assay: The cytotoxicity of samples on A549 cells was detected by MTT assay (Jiang and others 2012). Cells were seeded into 96-well plates and incubated with samples for 24, 48 or 72 h. Then the supernatant was removed and 100 μL of MTT solution was added to each well and incubated for by 4 h incubation at 37°C. The supernatant was sucked out and 200 μL of DMSO was added into each well. The amount of MTT formazan was quantified by measuring absorbance at 550 nm.
Semi-quantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR) analysis: A549 cells (1 × 106) were grown in 6-well plates for 24 h. Then cells were treated with sample with different time (0, 3, 6, 12, 24 and 48 h). Total RNA of the cells was isolated with a Trizol RNA isolation kit (Invitrogen, Carlsbad, CA, USA). The total RNA was reversetranscribed to cDNA and used as the template for PCR amplification. The forward and reverse primers were as follows: 5'-AGTGGAGGCCGACTTCTTGT-3' and 5'- CTGTTGCCACCTTTCGGTTA-3' for caspase-3, 5'- TGGTGGAGAACCCAAAGG-3' and 5'- GTCAAAGGAACCAAAGTCACG-3' for superoxide dismutase-2 (SOD-2), and 5'- TGTTACCAACTGGGACGACA-3' and 5'- CTCTCAGCTGTGGTGGTGAA-3' for β-actin. The amplified PCR products obtained by PCR were separated on 1% agarose gel electrophoresis, and the gel was stained with ethidium bromide. The gel was photographed with a Mini BIS Image Analysis System (DNR Bio-Imaging Systems Ltd., Jerusalem, Israel).
Statistical analysis
The data are represented as mean ± standard deviations of the experiments. A one-way analysis of variance (ANOVA) with post hoc test was performed to determine the differences between the groups using a commercially available program (SPSS 12 for Windows, SPSS Inc., Chicago, IL, USA). The level of significance was 0.05.
Results
Anti-inflammatory assay
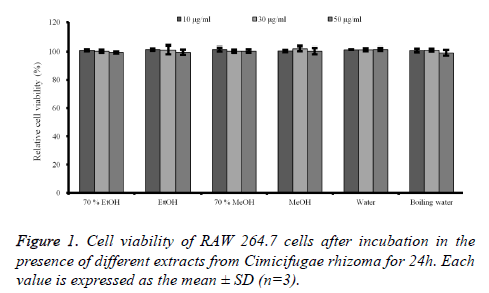
Cell cytotoxicity assay: The effects of C. rhizoma on RAW 264.7 cells were presented in figure 1. The results showed that no groups showed any statistically significant toxicity in the RAW 264.7 cells (P>0.05).
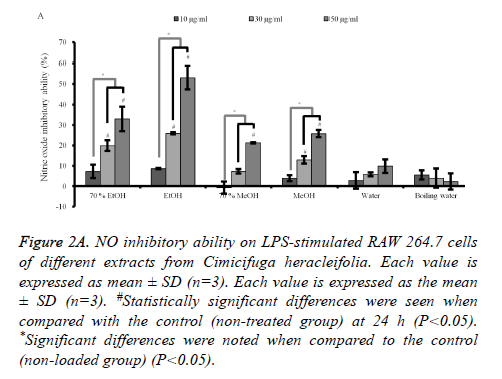
Quantification of NO production in LPS-induced RAW 264.7 cells: Stimulation with LPS for 24 h led to a robust increase in the NO production. However, C. rhizoma significantly suppressed NO by the LPS-stimulated RAW 264.7 cells in 70% ethanol, absolute ethanol, 70% methanol and absolute methanol groups with significant NO inhibition of 50 μg/ml concentration of each group of 32.9 ± 6.0 %, 53.0 ± 5.8 %, 21.2 ± 0.2 %, and 25.8 ± 1.8 %, respectively (Figure 2A). However, statistical significant decreases of NO production in 30 μg/ml concentrations were noted only in 70% ethanol, absolute ethanol, and absolute methanol with NO reduction of 19.8 ± 2.6 %, 25.9 ± 0.5 %, and 12.9 ± 1.8 %, respectively.
Figure 2A. NO inhibitory ability on LPS-stimulated RAW 264.7 cells of different extracts from Cimicifuga heracleifolia. Each value is expressed as mean ± SD (n=3). Each value is expressed as the mean ± SD (n=3). #Statistically significant differences were seen when compared with the control (non-treated group) at 24 h (P<0.05). *Significant differences were noted when compared to the control (non-loaded group) (P<0.05).
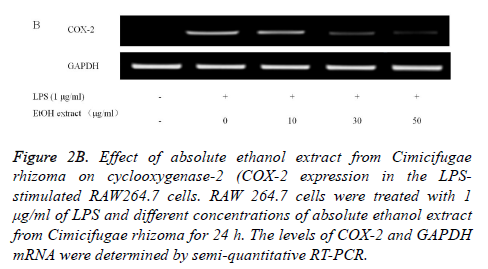
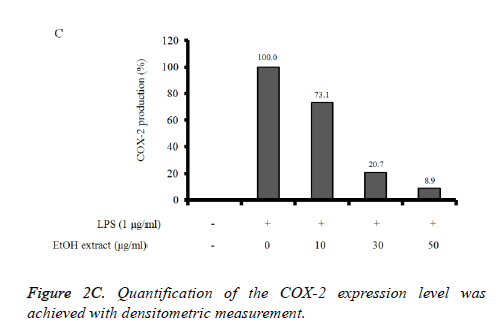
Reverse Transcription-Polymerase Chain Reaction (RTPCR) analysis: The COX-2 mRNA expression in the unstimulated RAW 264.7 cells was minimal, but expressions were profoundly induced after the treatment with LPS (Figure 2B). Pretreatment with absolute ethanol extract of C. rhizoma suppressed the LPS-stimulated COX-2 expression. The suppression of inflammatory related genes increased with increasing concentration of C. rhizoma extract (Figure 2C).
Figure 2B. Effect of absolute ethanol extract from Cimicifugae rhizoma on cyclooxygenase-2 (COX-2 expression in the LPSstimulated RAW264.7 cells. RAW 264.7 cells were treated with 1 μg/ml of LPS and different concentrations of absolute ethanol extract from Cimicifugae rhizoma for 24 h. The levels of COX-2 and GAPDH mRNA were determined by semi-quantitative RT-PCR.
Tumor suppressor activity assay
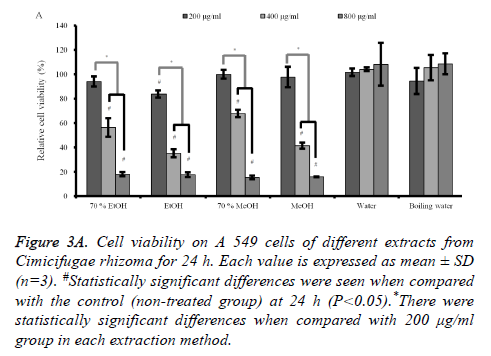
Cytotoxicity assay and semi-quantitative RT-PCR analysis: The results of cell cytotoxicity of A549 cells after incubation in the presence of different extracts from C. rhizoma for 24 h, 48 h and 72 h are shown in figures 3A-3C. The results at 24 h showed that C. rhizoma extract significantly reduced cellular viability when compared with untreated control in 70% ethanol, absolute ethanol, 70% methanol and absolute methanol groups (P<0.05) (Figure 3A).
Figure 3A. Cell viability on A 549 cells of different extracts from Cimicifugae rhizoma for 24 h. Each value is expressed as mean ± SD (n=3). #Statistically significant differences were seen when compared with the control (non-treated group) at 24 h (P<0.05).*There were statistically significant differences when compared with 200 μg/ml group in each extraction method.
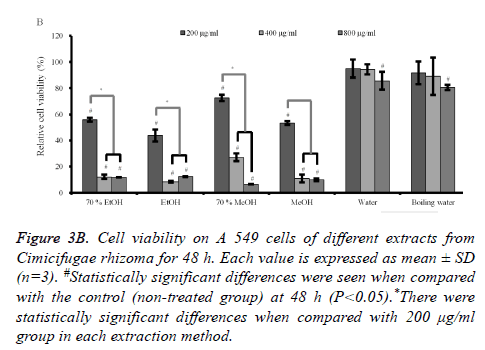
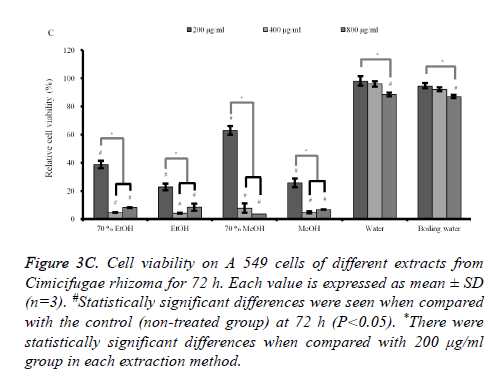
The results at 48h and 72 h (Figures 3B and 3C) showed similar trends with 24 h data with the decrease of viability with longer incubation time. The subgroups (from 400 to 800 μg/ml) of 70% ethanol, absolute ethanol, 70% methanol and methanol extracts showed statistically significant reduction in cellular viability with highest effects in absolute ethanol group.
Figure 3B. Cell viability on A 549 cells of different extracts from Cimicifugae rhizoma for 48 h. Each value is expressed as mean ± SD (n=3). #Statistically significant differences were seen when compared with the control (non-treated group) at 48 h (P<0.05).*There were statistically significant differences when compared with 200 μg/ml group in each extraction method.
Figure 3C. Cell viability on A 549 cells of different extracts from Cimicifugae rhizoma for 72 h. Each value is expressed as mean ± SD (n=3). #Statistically significant differences were seen when compared with the control (non-treated group) at 72 h (P<0.05). *There were statistically significant differences when compared with 200 μg/ml group in each extraction method.
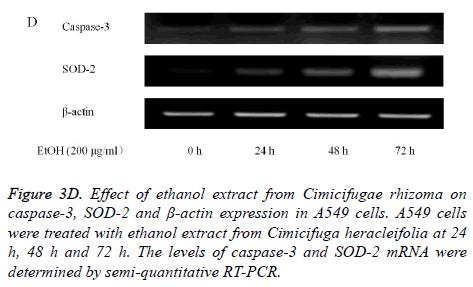
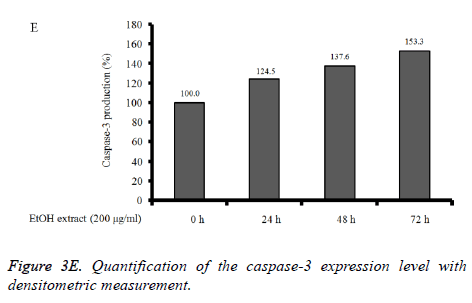
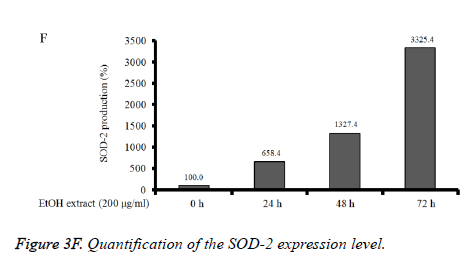
Absolute ethanol extracted most powerful effects of 22.8 ± 2.5 %, 4.1 ± 0.3 %, and 8.5 ± 2.5% with the concentration of 200, 400, and 800 μg/ml, respectively on 72 h when the control was considered 100 (100.0 ± 0.9) % (P<0.05). Water and boiling water extracts only showed significant effects at 48 h and 72 h with the concentration of 800 μg/ml. The expression of caspase-3 and SOD-2 increased with the increase of exposure time with C. rhizoma extracts (Figures 3D-3F).
Figure 3D. Effect of ethanol extract from Cimicifugae rhizoma on caspase-3, SOD-2 and β-actin expression in A549 cells. A549 cells were treated with ethanol extract from Cimicifuga heracleifolia at 24 h, 48 h and 72 h. The levels of caspase-3 and SOD-2 mRNA were determined by semi-quantitative RT-PCR.
Discussion
This result showed the effects of different extracts of C. rhizoma on the mouse leukaemic monocyte macrophage and lung carcinoma cell lines under predetermined concentrations. Absolute ethanol extracts showed highest anti-inflammatory and tumor suppressor activities on this experimental setting.
This study demonstrated the anti-inflammatory activities and highest effects were driven from the absolute ethanol of C. rhizoma from the LPS-activated macrophages. Fukinolic acid, fukiic acid, caffeic acid, ferulic acid, and isoferulic acid have been isolated and these components may produce antiinflammatory effects [10,11]. C. rhizoma was suggested to show inhibitory effects on histamine, bradykinin and COX-2 mediated inflammatory actions [5] and this studied confirmed the inhibition of NO production via reduction of COX-2 production.
Many anticancer drugs have been developed, the use of most of them is limited due to their toxicity to normal cells and tissues [12]. Our previous study showed that no toxicity was seen for the mesenchymal stem cells in 0.001-10μg/ml concentration up to day 3 [13,14]. The half maximal Inhibitory Concentration (IC50) values of C. rhizoma on primary cultured normal mouse hepatocyte were 80 μg/mL [8]. However, active constituents from Cimicifuga foetida promoted the proliferation for rat osteoblastoma cell line at the concentration of 0.001 μg/ml concentration and deoxyactein is reported to have the protective effects [15,16].
C. rhizoma extract showed cytotoxicity toward human cancer cell lines including promyelocytic cell line, lung carcinoma cell line, lung carcinoma cell line and human colon adenocarcinoma cell line [7]. The cytotoxicity of C. rhizoma was tested and the IC50 values on hepatocelluar carcinoma cell line, drug-resistant hepatocellular carcinoma cell line, and primary cultured normal mouse hepatocyte were 21, 43 and 80 μg/mL, respectively [8].
C. rhizoma extracts were reported to show tumor suppressor activities toward human tumor cell lines including promyelocytic leukemia, hepatocellular carcinoma, alveolar basal epithelial carcinoma, estrogen receptor positive breast carcinoma, primary colon carcinoma and myelogenous leukemia [7]. This study showed that the absolute ethanol extract showed highest tumor suppressor activities. Cimigenol, components isolated from C. rhizoma exerted the potent cytotoxic activity with IC50 values of 7.87 μM for hepatocellular carcinoma and 12.16 μM for alveolar basal epithelial carcinoma [7]. Activation of caspase-3 was related apoptosis and SOD-2 enzyme was considered a tumor suppressor [8,17] and changes in the transcriptional level of caspase-3 and SOD-2 may explain this phenomenon.
Within the limits of this study, C. rhizoma showed antiinflammatory effect using RAW 264.7 cell line and tumor suppressor activities on A549 cells and the effects were influenced by extraction methods. Absolute ethanol extract showed the highest anti-inflammatory and tumor suppressor activities in these experimental settings.
Acknowledgements
This study was partly supported by the 2015 Research Grant from Kangwon National University (No. 520150324), Republic of Korea and partly supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, Information and Communication Technology & Future Planning (NRF-2014R1A1A1003106), Republic of Korea. The authors report no conflicts of interest related to this study. The author does not have any financial interest in the companies whose materials are included in the article.
References
- Li JX, Yu ZY. Cimicifugae rhizoma: from origins, bioactive constituents to clinical outcomes. Curr Med Chem 2006; 13: 2927-2951.
- Li JX, Kadota S, Li HY, Miyahara T, Wu YW, Seto H. Effects of Cimicifugae rhizoma on serum calcium and phosphate levels in low calcium dietary rats and on bone mineral density in ovariectomized rats. Phytomedicine 1997; 3: 379-385.
- Sakurai N, Nagai M. Chemical constituents of original plants of Cimicifugae rhizoma in Chinese medicine. Yakugaku Zasshi 1996; 116: 850-865.
- Liao JF, Jan YM, Huang SY, Wang HH, Yu LL, Chen CF. Evaluation with receptor binding assay on the water extracts of ten CNS-active Chinese herbal drugs. Proc Natl Sci Counc Repub China B 1995; 19: 151-158.
- Kim SJ, Kim MS. Inhibitory effects of cimicifugae rhizoma extracts on histamine, bradykinin and COX-2 mediated inflammatory actions. Phytother Res 2000; 14: 596-600.
- Jainsu_New_Medical_College. Dictionary of Chinese Material Medica. Shanghai: Shanghai Scientific and Technological Publishing Co.; 1977.
- Lu L, Chen JC, Li Y, Qing C, Wang YY, Nian Y. Studies on the constituents of Cimicifuga foetida collected in Guizhou Province and their cytotoxic activities. Chem Pharm Bull (Tokyo) 2012; 60: 571-577.
- Tian Z, Pan R, Chang Q, Si J, Xiao P, Wu E. Cimicifuga foetida extract inhibits proliferation of hepatocellular cells via induction of cell cycle arrest and apoptosis. J Ethnopharmacol 2007; 114: 227-233.
- Jiang Y, Hu W, Han W, Yeo JH, Wang MH. Antioxidant and nitric oxide production inhibitory activities of scouring rush (Equisetum hyemale L.). Food Sci Biotechnol 2012; 21: 1037-1044.
- Takahira M, Kusano A, Shibano M, Kusano G, Sakurai N, Nagai M. Three new fukiic acid esters, Cimicifugic acids A, B and C, from Cimicifuga simplex WORMSK. Chem Pharm Bull 1998; 46: 362-365.
- Hasa Y, Tazaki H. Biosynthesis of fukinolic acid isolated from Petasites japonicus. Biosci Biotechnol Biochem 2004; 68: 2212-2214.
- Liu L, Wang MH. Cloning, expression, and characterization of a methionine sulfoxide reductase B gene from Nicotiana tabacum. Protein J 2013; 32: 543-550.
- Jeong SH, Kim BB, Lee JE, Ko Y, Park JB. Evaluation of the effects of Angelicae dahuricae radix on the morphology and viability of mesenchymal stem cells. Mol Med Reports 2015; 12: 1556-1560.
- Jeong SH, Lee JE, Kim BB, Ko Y, Park JB. Effects of Cimicifugae Rhizoma on the morphology and viability of mesenchymal stem cells. Exp Ther Med 2015; 10: 629-634.
- Zhao XH, Chen DH, Si JY, Pan RL, Shen LG, Chen D. Studies on new trierpenoid constituents from the Rhizoma of Cimicifuga foetida. Zhongguo Zhong Yao Za Zhi 2003; 28: 135-138.
- Choi EM. Deoxyactein Isolated from Cimicifuga racemosa protects osteoblastic MC3T3-E1 cells against antimycin A-induced cytotoxicity. J Appl Toxicol 2013; 33: 488-494.
- Bravard A, Sabatier L, Hoffschir F, Ricoul M, Luccioni C, Dutrillaux B. SOD2: a new type of tumor-suppressor gene? Int J Cancer 1992; 51: 476-480.