Short Communication - Biomedical Research (2019) Volume 30, Issue 2
Analyses of single nucleotide polymorphisms in Angiostrongylus cantonensis based on transcriptome sequencing.
Liang Yu1,2, Wenzhen Fang3 and Damin Luo1,2*
1Department of Biology, School of Life Sciences, Xiamen University, P.R. China
2State Key Laboratory of Cellular Stress Biology, Xiamen University, P.R. China
3College of the Environment & Ecology, Xiamen University, P.R. China
- *Corresponding Author:
- Damin Luo
Department of Biology, School of Life Sciences
Xiamen University, P.R. China
Accepted date: March 25, 2019
DOI: 10.35841/biomedicalresearch.30-19-134
Visit for more related articles at Biomedical ResearchAbstract
Objective: This study aimed to reveal the SNP characters of Angiostrongylus cantonensis based on transcriptome sequencing for further using. Method: SAMtools and GATK2 were used for SNP detection. SNP frequency and transition verse transversion ratio were calculated. SNPs containing unigenes were annotated with six data bases. Results: A total number of 71,214 SNPs were predicted based on the transcriptomes of the fifth stage larvae (L5) and female adults (F). 53,019 of them were predicted as non-synonymous while another 18,195 were predicted as synonymous SNPs. Statistical analysis of all these predicted SNPs indicated that the estimated SNP frequency was 0.21% (one SNP per 476 bp) and the estimated ratio for transition to transversion was 2.125. Moreover, gene function analysis including GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway enrichment analysis of all SNPs containing unigenes were conducted for the better understanding of their functions and significances in genetics. Conclusion: The results will help us to understand the basic characters of A. cantonensis SNPs fully and the predicted SNPs will be powerful tools for species authentication development, transmission survey, as well as many other genome wide association studies (GWAS).
Keywords
Angiostrongylus cantonensis, Transcriptome, SNP characters
Introduction
A. cantonensis is a well-known food-borne causative etiological agent of human eosinophilic meningoencephalitis. It is now epidemic in a very wide region of the world, including Asia, the Pacific Islands, the Caribbean islands and Brazil. During the past decade, more than 2,800 cases of human infections and several human angiostrongyliasis outbreaks have been reported and which declared it as a public health problem that cannot be neglected [1].
Single nucleotide polymorphism (SNP) markers are useful in species authentication, transmission investigation and evolution researches besides GWAS in parasite [2]. Transcriptome sequencing has become the major efficient and cost-effective method for rapid SNP discovery of the expressed genes even in non-model species. Pool samples from different individuals for high throughput transcriptome sequencing are efficient strategy to infer genetic variation in a population and which have been widely proofed [3]. In the present study, SNPs were predicted by analysis of reads from two transcriptomes of A. cantonensis and further researched their characterization.
Materials and Methods
Basial data information
L5 and F total RNA were sequenced on an Illumina HiSeq 2500 platform using the paired-end RNA-Seq method. Clean sequencing data from these two libraries were pooled together for a global transcriptome de novo assembly by Trinity (v2.0.6). This comprehensive transcriptome was defined as P transcriptome. Then, transcriptome of each stages was assembled and the P transcriptome was used as reference in our previous research [4].
SNPs detection, annotation and statistics
Indexed pair-end clean reads of each sample and mapping them to the P transcriptome with BAM. Then, Picard tools (version 1.41) and SAMtools (version 0.1.18) were used to sort, remove duplicated reads and merge the BAM alignment results of each sample. GATK2 software was utilized to perform SNP calling. Raw vcf files were filtered with GATK standard filter method and only SNPs with distance larger than 5 were retained. In order to obtain more reliable SNPs, only those with quality score over 30 and the reads depth over 5 were approbatory as predicted SNPs. SNP frequency among transcriptomes was calculated by dividing the total length of reference by the total number of SNPs. Transition verse transversion ratio was derived by analyzing each type of DNA substitution.
Transcriptome annotation was performed with a conventional procedure. Briefly, the unigenes were annotated with BLASTx (BLAST+v2.2.25) by querying them to the following databases: NCBI non-redundant protein sequences (Nr), NCBI non-redundant nucleotide sequences (Nt), the Protein Family database (Pfam), Swiss-Prot, Gene Ontology (GO), the eukaryotic orthologous groups database (KOG) and KEGG. The E-value cutoff was set as 1 × 10-5.
Results
50,234,050 and 55,725,337 raw reads were generated from L5 and F transcriptome Illumina sequencings, respectively. After filtered with QC Toolkit, 101,233,882 high-quality reads were obtained for P transcriptome assembly. Eventually, 51,401,554 unigenes with an average length of 621 bp were produced in the P transcriptome.
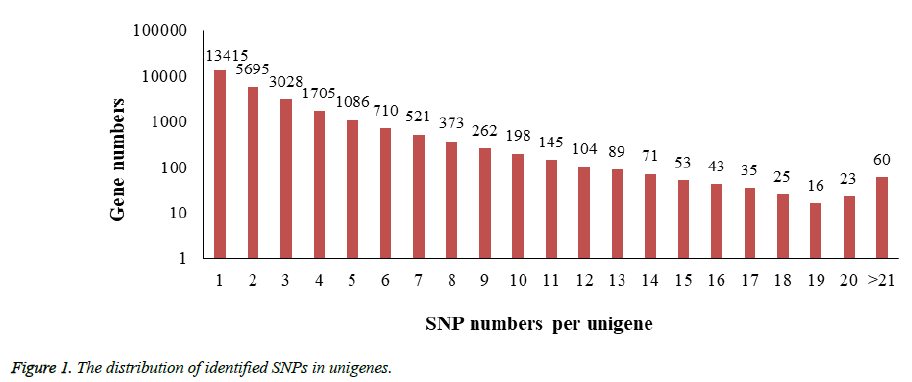
In the process of SNPs detection, a total number of 71,214 SNPs distributed in 27,657 unigenes (33.4% of all unigenes) were identified and the estimated SNP frequency was 0.23% (one per 429 bp). 48.5% of unigenes contain only a single SNP while another 664 unigenes possess more than 10 SNPs in each one. Figure 1 revealed the details of the SNPs distribution among those unigenes.
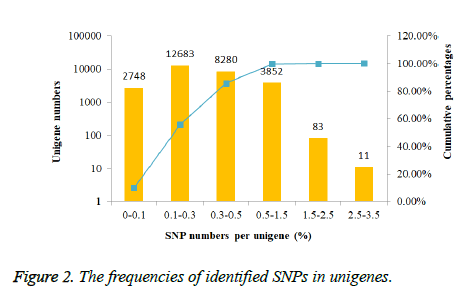
The frequencies of identified SNPs in each unigene were calculated by dividing unigene length by SNPs number per unigene.
In order to investigate the mutation rate among unigenes, the SNP frequency within unigenes was reckoned. According to the results showed in Figure 2, nearly half unigenes SNP frequencies were in the range from 0.1% to 0.3%. However, only 94 unigenes had the SNP frequencies bigger than 1.5%.
Among these SNPs, more transitions substitution (68%) were discovered than transversion substitution (32%) (Table 1). In terms of transition substitution, the amount of A/G transitions was similar to that of C/T transition. As for transversion substitution, the frequency of A/T was slightly higher than other types (G/C, G/T, A/C). In addition, the estimated ratio for the transition to transversion was 2.125.
| Type | Transition | Transversion | ||||
|---|---|---|---|---|---|---|
| G/A | C/T | G/C | G/T | A/C | A/T | |
| Numbers | 23952 | 24490 | 5708 | 5180 | 5380 | 6504 |
| Percentage | 0.336 | 0.344 | 0.08 | 0.073 | 0.076 | 0.091 |
Table 1: Statistics of transition and transversion type in total SNPs.
In addition, all SNPs were classified into two categories which including coding SNPs and non-coding SNPs. Moreover, the coding SNPs were divided into two subgroups according to whether the encoded amino acid changed or not. Put simply, 18,195 SNPs were located in the open reading frame (ORF) including 8,095 non-synonymous SNPs and 10,100 synonymous SNPs, another 53,019 SNPs were located in UTR (untranslated region). Furthermore, all these non-synonymous SNPs were distributed in 4,458 unigenes while synonymous SNPs were distributed in 6,694 unigenes.
After unigenes function annotation, 10,159 (36.7%) of 27,657 SNPs containing unigenes had significant hit to the proteins in the non-redundant (Nr) database, and 7,862 (28.4%) unigenes were assigned with one or more GO ID. Unigenes in ‘binding’, ‘cellular processes’ and ‘cell’ are the dominated terms of each subclass. On the other hand, 4,901 unigenes (17.7%) with 22,566 SNPs were allocated to KOG 25 terms, and most of them were genes associated with the “Signal transduction mechanisms”. KEGG analysis results demonstrated that 3,229 (11.7% of total) unigenes containing 14,650 SNPs could be annotated with the KEGG database. And these unigenes could be assigned to 297 KEGG pathways. The top 10 KEGG pathways were displayed in Table 2. As shown in the table, lysine degradation pathway is the largest group one of them, followed by purine metabolism and protein processing in endoplasmic reticulum pathways.
| KEGG pathways | Unigene numbers |
|---|---|
| Lysine degradation | 107 |
| Purine metabolism | 84 |
| Protein processing in endoplasmic reticulum | 82 |
| Spliceosome | 80 |
| Endocytosis | 78 |
| Oxidative phosphorylation | 75 |
| RNA transport | 74 |
| Carbon metabolism | 73 |
| Regulation of actin cytoskeleton | 73 |
| Ribosome | 73 |
Table 2: The top 10 most abundant KEGG pathways.
Discussion
An SNP marker is just a single base change in a DNA sequence with an alternative of two possible nucleotides at a given position, usually defined as transition or transversion. Some of they can provide valuable information on associations between specific genes or other DNA structures and phenotypes, or on population and genome dynamics. In principle, the ratio of transitions to transversions should be 2 if mutations are random. However, large amount of observed data indicates a distinct bias towards the transitions and this may mainly due to the high spontaneous rate of deamination of 5- methyl cytosine (5mC) to thymidine in the CpG dinucleotides [5]. The ratio in our research is 2.125, which is in accordance with this fact.
SNPs can alter protein function and phenotype. 8,095 of 18,198 SNPs were non-synonymous SNPs, which indicted that their corresponding coding amino acid would be changed, since this will be the more likely responsible for phenotypic variation. Another 53,019 SNPs were located in UTR, including exons, promoters or other regulatory regions, have the potential to affect the regulation of genes expression. Both of them could be the key suspects responsible for drug resistance or different virulence of A. cantonensis.
SNPs are the most useful and widely applied markers in molecular genetic studies. As a zoonotic parasite, A. cantonensis can infect many other kinds of wild animals besides human, even in many rare species of the world, such as Ammotragus lervia, Macrotis lagotis and Diplothrix legata [6]. Parasites usually detriment the host organism by imposing high costs on populations, increasing morbidity and mortality, particularly in the hosts under ecological stress [7]. Therefore, it is crucial to hunt out the route of transmission, then implement corrective and preventive measures for re-wilding, and protect project of rare animals.
Conclusion
This work is the first effort towards developing SNP markers in A. cantonensis. And 71,214 high-quality SNPs were successfully predicted. The estimated SNP frequency was 0.21% (one SNP per 476 bp) and the estimated ratio for the transition to transversion was 2.125. These SNPs applied for the development of molecular markers to distinguish different parasite strains will be very helpful for primary infection source survey and which in turns will be favorable for prevention strategy development. Furthermore, the predicted SNPs may also provide a fruitful approach towards the antihelmintic drugs and vaccines development in further studies.
Acknowledgement
This research was supported by the National Sharing Service Platform for Parasite Resource (TDRC-22).
References
- Tunholi-Alves VM, Tunholi VM, Gôlo P, Lima M, Garcia J, Júnior AM, Pontes EG, Bittencourt VR, Pinheiro J. Effects of infection by larvae of Angiostrongylus cantonensis (Nematoda, Metastrongylidae) on the lipid metabolism of the experimental intermediate host Biomphalaria glabrata (Mollusca: Gastropoda). Parasitol Res 2013; 112:2111-2116.
- Van Paridon BJ, Goater CP, Gilleard JS, Criscione CD. Characterization of nine microsatellite loci for Dicrocoelium dendriticum, an emerging liver fluke of ungulates in North America, and their use to detect clonemates and random mating. Mol Biochem Parasitol 2016; 207:19-22.
- Raineri E, Ferretti L, Esteve-Codina A, Nevado B, Heath S, Perez Enciso M. SNP calling by sequencing pooled sample. BMC Bioinformatics 2012; 13:239.
- Yu L, Cao B, Long Y, Tukayo M, Feng C, Fang W, Luo D. Comparative transcriptomic analysis of two important life stages of Angiostrongylus cantonensis: fifth-stage larvae and female adults. Genet Mol Biol 2017; 40:540-549.
- Vignal A, Milan D, Sancristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol 2002; 34:275-305.
- Spratt DM. Species of Angiostrongylus (Nematoda: Metastrongyloidea) in wildlife: A review. Int J Parasitol Parasites Wildl 2015; 4:178-189.
- Clough D, Kappeler PM, Walter L. Genetic regulation of parasite infection: empirical evidence of the functional significance of an IL4‐gene SNP on nematode infections in wild primates. Front Zool 2011; 8:9.