Research Article - Journal of Molecular Oncology Research (2018) Volume 2, Issue 3
SCCAg mRNA expression predicts early relapse in histopathologically negative lymphnodes of oral squamous cell carcinoma patients.
Trupti Trivedi1*, Rajan Tankshali2, Dhaval Jetly3, Nandita Ghosh4 and Prabhudas Patel5
1Clinical Carcinogenesis Lab-3, Gujarat Cancer and Research Institute, India
2Surgical Oncology Department, Gujarat Cancer and Research Institute, India
3Pathology Department, Gujarat Cancer and Research Institute, India
4Tumor Biology Lab-2, Gujarat Cancer and Research Institute, India
5Cancer Biology Department, Gujarat Cancer and Research Institute, India
- *Corresponding Author:
- Dr. Trupti Trivedi
Clinical Carcinogenesis Lab-3
Gujarat Cancer & Research Institute
Gujarat, India
E-mail: truptitrivedi99@yahoo.ca
Accepted date: August 28, 2018
Citation: Trivedi T, Tankshali R, Jetly D et al.. SCCAg mRNA expression predicts early relapse in histopathologically negative lymphnodes of oral squamous cell carcinoma patients. J Mol Oncol Res. 2018;2(3):48-58.
DOI: 10.35841/molecular-oncology.2.3.48-57
Visit for more related articles at Journal of Molecular Oncology ResearchAbstract
Local recurrence and lymphnode (LN) metastasis are the most significant risk factors for morbidity and mortality for oral squamous cell carcinoma (OSCC) patients. In the current study, we aimed to evaluate the clinical significance of SCCAg mRNA expression in OSCC patients having histopathologically negative lymphnodes (HNLNs) and pretherapeutic peripheral blood samples (PPBs.). SCCAg mRNA was evaluated in total N=123 samples using nested RT-PCR technique. SPSS statistical software was used and p value <0.05 was considered as significant. The analysis revealed that in HNLNs, the mean ? SE for SCCAg mRNA expression was 1794.98 ? 106.67 with a median of 1488.0 while in PPBS it was 2308.27 ? 196.10 with a median of 1985.0. The frequency of SCCAg mRNA in HNLNs and in PPBs was 27% (15/55) and 29% (8/28), respectively. SCCAg mRNA expression in HNLNs and in PPBs showed a significant positive correlation with tumor size and lymphatic permeation. Multivariate survival analysis for RFS demonstrated that only SCCAg mRNA expression in HNLNs (p=0.001) and PPBs (p=0.001) were the most significant independent prognostic factors. However, for OS, multivariate analysis showed that SCCAg mRNA of HNLNs emerged at step 2 after tumor size and for PPBs it remained as single most significant independent prognostic marker. Thus, SCCAg mRNA may represent a useful tool for more accurate staging, which could improve disease management and help to obtain maximal therapeutic benefit from adjuvant therapies. Hence, SCCAg mRNA transcript could serve as a useful independent predictor of disease relapse, suggesting an increased risk of silent metastasis.
Keywords
SCCAg mRNA, Histopathologically N0 nodes, Oral squamous cell carcinoma, Multivariate analysis, Prognostic significance
Introduction
Today the world is heading towards "modern epidemics” in which various non-communicable diseases including cancer are emerging as major public health problems. Amongst all cancers, head and neck represents the seventh most common malignancy, worldwide [1], amongst, oral squamous cell carcinoma (OSCC) has the highest incidence of all head and neck squamous cell carcinoma cancers (HNSCCs) [2]. The incidence of OSCC varies markedly by geographic region, and more than half of all cancer cases occur in developing countries [3]. India has one of the highest incidences of oral cancer and accounts for about 30% of all new cases annually [4]. OSCC is characterized by an aggressive growth pattern, high degree of local invasiveness and cervical lymph node spread. The presence of regional neck metastases is a major determinant of both prognosis and treatment decisions in patients with OSCC [5]. However, the low sensitivity of currently available diagnostic modalities is a problem, because a fairly high proportion (30%-40%) of lymph node metastasis (LNM) is left undetected in this population. Lymph node metastasis can be an early event in disease progression and is associated with a 50% decrease in survival, being one of the most important independent prognostic factors. Thus, for clinically node negative neck OSCC patients the dilemma in current clinical management of the neck is the choice between possible under-treatment of 30%-40% of patients with occult metastases in patients with OSCC, would benefit greatly from staging techniques that add accuracy to the assessment of nodal disease. Hence, sensitive methods to evaluate the status and prognosis of these patients with nodal micrometastasis are desirable [6].
Until recently, serial sectioning and immunohistochemistry of cervical LNs were suggested for detection of micrometastasis. Using conventional [7,8] or serial sectioning with haematoxylin and eosin (H and E) staining [9,10] only rarely have occult metastatic tumor cells been detected. However, staining with epithelial specific antibodies has revealed the presence of occult metastatic tumor cells in LNs of HNSCC [11-13]. In this regard, Ambrosch et al. have reported an incidence of 7.9% in the neck dissection specimens from HNSCC patients who were originally staged as pathologically N0. Further, Enepekides et al. found a 5% incidence in pathologically No patients with T1 or T2 SCC of the lip and oral cavity. Thus, these data indicated that serial sectioning and immunohistochemical assessment of cervical LNs can be useful to refine the staging system and a significant percentage of cases found to have micrometastasis would be upstaged. However, the prognostic significance of these findings remains unclear, also, for immunohistochemistry, serial microscopic sectioning of the nodes may reveal micrometastases, but is impractical for routine use. Recently, there has been considerable progress in molecular diagnostics in these areas.
Detection of micrometastasis in regional LNs by RT-PCR has been reported in HNSCC [14-16]. However, none of these investigating groups have evaluated the prognostic value of these markers in HNSCC. Also, in HNSCC, there are very few reports that have shown disseminated micrometastatic cells in blood [17-19]. Due to lack of specificity of keratins, several authors have suggested ck20 to be an inappropriate marker for detecting micrometastasis [20]. Brakenhoff et al. by applying E48, demonstrated micrometastatic cells in 10% of the blood samples of patients with HNSCC. Again the relationship between presence of E48 in blood and prognosis was not evaluated. Hence, in OSCC, detection of micrometastasis in histopathologically negative lymph nodes (HNLNs) and pretherapeutic peripheral blood samples (PPBs) is of significance as presence of LN metastasis is still the most significant prognostic factor that determines clinical management. Therefore, in the present study we examined the importance of SCCAg mRNA transcript as a marker to detect micrometastasis in HNLNs and PPBs of patients with oral cancer with following specific aims: (i) to detect SCCAg mRNA in HNLNs and PPBs of patients with OSCC and thereby identify high-risk group of patients, (ii) to explore the relation of SCCAg mRNA level of HNLNs and PPBs with the clinicopathological parameters of primary tumors and (iii) to evaluate the prognostic significance of SCCAg mRNA expression in HNLNs and PPBs of patients with OSCC.
Materials and Methods
Patients
SSCAg mRNA was studied in total N=123 samples using nested RT-PCR technique. Apart from HNLNs (N=55) and PPBs from OSCC patients (N=28), reference control group of N=40 (N=20; metastatic LNs of breast and colorectal cancer, N=10; primary tumors of OSCC, N=10; peripheral blood from healthy individuals) were included in the current study. None of the OSCC patients received any therapies prior to surgery. Pretherapeutic blood samples and surgical specimens of included patients were collected from Department of Surgical Oncology, GCRI, Civil Hospital Compound, Ahmedabad, India. The median age of patients included in the study was 45 years with the range of 25-75 years. Majority of patients (71%) of patients had habit of tobacco and/or smoking. The disease was staged according to the criteria of the International Union Against Cancer pTNM classification (UICC). In the present study out of 55 patients, 38 patients (69%) had T1-T2 tumors, whereas, 31% of patients had T3-T4 tumors (Table 1). Forty-five percent of patients had well differentiated tumors and 55% patients had moderate or poorly differentiated tumors. Further, 81% and 71% of patients showed absent of lymphatic and vascular permeation, respectively (Table 1). Tissue specimens were collected directly from the operation theatre, on ice. A portion of negative lymph node was selected by a pathologist, and was as snap frozen in liquid nitrogen and preserved at −85°C for total RNA extraction. Total RNA were extracted from the peripheral blood and negative lymphnodes after confirmed by pathological report. Extracted total RNA were preserved at −85°C till further analysis.
| Primers | Position | Primer sequences | bp* |
|---|---|---|---|
| Outer primers | |||
| A | 7 | 5'-GCC CAC CTC TGC TTC CTC TA-3' | 1083 |
| B | 1070 | 5'-GCT TCT GCT CCC TCC TCT GT-3' | |
| Inner primers | |||
| C | 470 | 5'-GCA AAT GCT CCA GAA GAA AG-3' | 61 |
| D | 711 | 5'-CGA GGC AAA ATG AAA AGA TG-3' |
Table 1: Primers used for SCCAg mRNA expression.
Methodology
Total RNA from all samples was extracted according to the method of Chomczynski et al. For SCCAg mRNA expression, nested RT-PCR was performed and the amplified products run on 1.5% agarose gels were semiquantitated on gel scan densitometer.
Nested RT-PCR
Nested RT-PCR is a technique used for sensitive detection of gene expression. First RT-PCR followed by nested PCR was performed for sensitive gene detection. The first round RT-PCR was performed using the GeneAmp EZ rTth RNA PCR kit (Perkin Elmer, USA). A master mix was prepared by adding the reagents in the proportion shown below: 5X EZ Buffer; 10.0 μl, 10 mM dNTP 1.5 μl each, rTth DNA polymerase 2.0 μl, Mn(OAc) solution; 5 μl, sense and antisense primers (The outer and inner primers used for first round RT-PCR followed by nested PCR are depicted in Table 1), 5.0 μl each, 1 μg total RNA for total volume of 50 μl reaction. The sequences of the outer and inner primers were similar to those used by Kano et al. and were designed from the published sequence of human SCCAg cDNA (Suminami et al.).
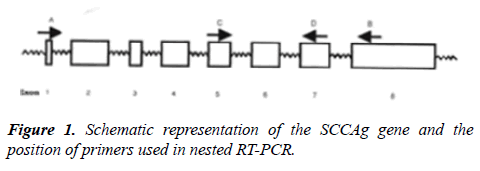
Primers; Stock solution-1 μg/μl, working solution-20 ng/μl (Sigma Genosys Ltd) were used. To avoid false positive results caused by genomic DNA contamination, primers were selected which spanned introns (Figure 1). For HNLNs and PPBs, 1 μg of total RNA was amplified by RT-PCR. A master mix for RT-PCR was prepared by adding the reagents in the proportions and condition were as follows: Reverse transcription at 60°C for 30 min, Initial denaturation at 94°C for 1 min, Thirty-five cycles of PCR: 94°C for 45 sec, 51°C for 45 sec, 72°C for 30 sec, Final extension at 72°C for 7 min. With 3 μl of the first round RT-PCR amplified product, the second round of PCR was performed, using Taq DNA polymerase kit from Fermentas, Life Science. Taq DNA polymerase 5 U/μl in 10X PCR buffer, pH 8.8, 25 mM magnesium chloride, 2 mM dNTPs, 10X PCR Buffer, 5.0 μl, dNTPs, DNA polymerase; 0.25 μl, MgCl2 solution, Primers 5.0 μl each, First round product 3.0 μl, The master mix was equally distributed into 200 μl MicroAmp reaction tubes (Perkin Elmer, USA). Thereafter, 3 μl RT-PCR amplified product was pipetted into the respective tubes. PCR profile was as follows: Initial denaturation at 94°C for 1 min. Thirty-five cycles of PCR: 94°C for 45 sec, 59°C for 45 sec, 72°C for 1 min. Final extension at 72°C for 7 min. The PCR amplification product was electrophoresed on a 2.0% agarose gel containing ethidium bromide along with a 100 bp DNA ladder to confirm the 261 bp size of the amplified product. The intensity of PCR products were measured and integrated on Gel Documentation System (Bio-Rad) using the Molecular Analyst Software, in the units of counts/mm2. Positive controls included total RNA from primary tumor tissues obtained from OSCC patients.
Negative controls included total RNA from metastatic lymph nodes obtained from breast and colorectal cancer patients, Negative control for the nested round, consisted not addition of first round RT-PCR product of the negative control. Internal positive control used was GAPDH mRNA amplification, as described earlier.
Survival analysis
Survival analysis for SCCAg mRNA in HNLNs was performed in 76% (42/55) of OSCC patients, while in PPBs it was carried out in 86% (24/28) of patients, which could be followed for a period of 2 years. Relapse-free survival (RFS) and OS was evaluated using Kaplan-Meier survival estimation. Significant results observed were further analyzed in univariate and multivariate survival analysis to identify factors with an independent effect on risk of recurrence or death from oral cancer, using Cox proportional hazard regression model.
Statistical analysis
Statistical analysis was performed using the SPSS statistical software programme (release 19; Chicago, IL, USA, 1999). Logistic regression was carried out to determine significant marker alterations in different stages of oral cancer development. Two tailed chi-squared test was used to assess the association between clinicopathological parameters. Correlation between two parameters was calculated using Spearman’s correlation co-efficient (r) method. RFS and OS were calculated according to Kaplan-Meier survival estimation and significance was assessed using the Log rank test. Multivariate analysis was done using Cox forward step-wise proportional hazard regression model. The Wald statistic and odds ratio (OR) with 95% confidence interval (CI) were used to assess prognostic significance for RFS, OS and molecular alterations. Non parametric Mann-Whitney U test and paired ttest was carried out for analysis of mRNA transcript level and p53 antibody reactivity. p values ≤ 0.05 were considered to be significant.
Results
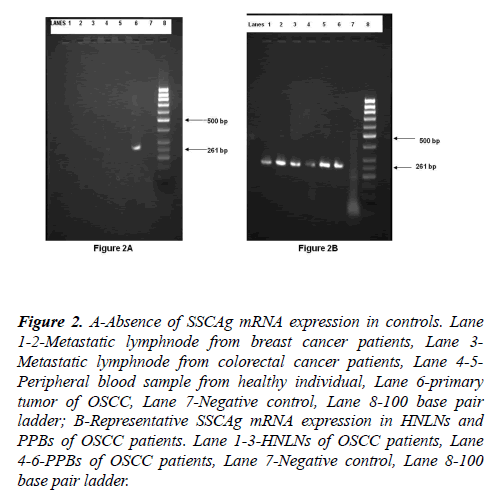
SCCAg mRNA expression was not detected in 20 metastatic LNs collected from breast and colorectal cancer patients or in 10 peripheral blood samples collected from healthy individuals (Figure 2). On the other hand, SCCAg mRNA was detected in all 10 primary tumors of patients with OSCC.
Figure 2: A-Absence of SSCAg mRNA expression in controls. Lane 1-2-Metastatic lymphnode from breast cancer patients, Lane 3- Metastatic lymphnode from colorectal cancer patients, Lane 4-5- Peripheral blood sample from healthy individual, Lane 6-primary tumor of OSCC, Lane 7-Negative control, Lane 8-100 base pair ladder; B-Representative SSCAg mRNA expression in HNLNs and PPBs of OSCC patients. Lane 1-3-HNLNs of OSCC patients, Lane 4-6-PPBs of OSCC patients, Lane 7-Negative control, Lane 8-100 base pair ladder.
SCCAg mRNA expression in HNLNs and PPBs of patients with OSCC
SCCAg mRNA expression in HNLNs and PPBs of patients with OSCC is shown in Table 2. Semi-quantitative analysis of SCCAg mRNA expression was performed to detect occult tumor cells in HNLNs (N=55) and PPBs (N=28) of patients with OSCC. The analysis revealed that in HNLNs, the mean ± SE for SCCAg mRNA expression was 1794.98 ± 106.67 with a median of 1488.0 while in PPBS it was 2308.27 ± 196.10 with a median of 1985.0. The mean ± SE was used as a cut-off for further analysis of the data. According to the cut-off value, the frequency of SCCAg mRNA in HNLNs and in PPBs was 27% (15/55) and 29% (8/28), respectively (Table 2).
| HNLNs (N=55) | PPBs (N=28) | |
|---|---|---|
| Range | 613.0-510.0 | 1196.0-5736.0 |
| Mean ± SE | 1794.98 ± 106.67 | 2308.21 ± 196.10 |
| Median | 1488 | 1985 |
| Cut-off value | 1901.65 | 2504.31 |
| Frequency of SCCAg expression | 27% | 29% |
Table 2: SCCAg mRNA expression in HNLNs and PPBs of patients with OSCC-Mann-Whitney U test.
Relation of SCCAg mRNA with clinicopathological features
The relation of SCCAg mRNA expression in HNLNs and PPBs with clinicoapathological features is depicted in Tables 3A and 3B, respectively.
| SCCAg mRNA | |||||
|---|---|---|---|---|---|
| Characteristics | N | <1901.65 N (%) | >1901.65 N (%) | r | p value |
| Total patients | 55 | 40 (73) | 15 (27) | ||
| Age (years) | 0.261 | 0.053 | |||
| ≤ 45 | 30 | 25 (83) | 05 (17) | ||
| >45 | 25 | 15 (60) | 10 (40) | ||
| Gender | 0.236 | NS | |||
| Male | 42 | 33 (79) | 09 (21) | ||
| Female | 13 | 07 (54) | 06 (46) | ||
| Family history | 0.212 | NS | |||
| Absent | 52 | 39 (75) | 13 (25) | ||
| Present | 3 | 01 (33) | 02 (67) | ||
| Anatomic site | 0.193 | NS | |||
| Buccal mucosa | 27 | 22 (81) | 05 (19) | ||
| Tongue | 28 | 18 (64) | 10 (36) | ||
| Tobacco habit | -0.07 | NS | |||
| Absent | 12 | 08 (67) | 04 (33) | ||
| Present | 43 | 32 (74) | 11 (26) | ||
Table 3A: Relationship between SCCAg mRNA expression and patient characteristics (HNLNs). NS: Not significant.
| SCCAg mRNA | |||||
|---|---|---|---|---|---|
| Characteristics | N | <1901.65 N (%) |
>1901.65 N (%) |
r | p value |
| Total patients | 55 | 40 (73) | 15 (27) | ||
| Tumor size | 0.385 | 0.004 | |||
| T1/T2 | 38 | 32 (84) | 06 (16) | ||
| T3/T4 | 17 | 08 (47) | 09 (53) | ||
| HG# | 0.067 | NS | |||
| I* | 25 | 19 (76) | 06 (24) | ||
| II** + III*** | 30 | 21 (70) | 09 (30) | ||
| Keratin | 0.171 | NS | |||
| Absent | 4 | 04 (100) | 00 (00) | ||
| Present | 51 | 36 (71) | 15 (29) | ||
| Growth pattern | -0.05 | NS | |||
| Ulcerative | 46 | 33 (72) | 13 (28) | ||
| Exophytic | 9 | 07 (78) | 02 (22) | ||
| LP@ | 0.332 | 0.014 | |||
| Absent | 42 | 34 (81) | 08 (19) | ||
| Present | 13 | 06 (46) | 07 (54) | ||
| VP$ | 0.099 | NS | |||
| Absent | 53 | 39 (74) | 14 (26) | ||
| Present | 2 | 01 (50) | 01 (50) | ||
| Notations: #Histological grade, @Lymphatic permeation, $Vascular permeation, *Well differentiated, **Moderately differentiated, ***Poorly differentiated, NS: Not significant. | |||||
Table 3B: Relationship between SCCAg mRNA expression and tumor characteristics (HNLNs).
Histopathological negative lymph nodes (HNLNs)
SCCAg mRNA expression in HNLNs when correlated with patient characteristics showed a trend towards significance with age only (r=+0.261, p=0.053) (Table 3A).
Likewise, SCCAg mRNA expression in HNLNs when associated with tumor characteristics showed a significant positive correlation with tumor size (r=+0.385, p=0.004) and lymphatic permeation (r=+0.332, p=0.014). However, statistical significance was not observed with rest of the tumor characteristics (Table 3B).
Pretherapeutic peripheral blood samples (PPBs)
The relation of SCCAg mRNA expression in PPBs with patient and tumor characteristics is depicted in Tables 4A and 4B, respectively. Significant correlation was not observed between SCCAg mRNA expression in PPBs and patient characteristics (Table 4A). However, SCCAg mRNA expression in PPBs when related with tumour characteristics showed a significant correlation with tumour size (r=+0.388, p=0.040) and lymphatic permeation (r=+0.388, p=0.040). Significant correlation was not observed with rest of the tumour characteristics (Table 4B).
| SCCAg mRNA | |||||
|---|---|---|---|---|---|
| Characteristics | N | <2504.31 N (%) |
>2504.31 N (%) |
r | p value |
| Total patients | 28 | 20 (71) | 08 (29) | ||
| Age (years) | 0.139 | NS | |||
| ≤ 45 | 17 | 13 (76) | 04 (24) | ||
| >45 | 11 | 07 (64) | 04 (36) | ||
| Gender | 0.088 | NS | |||
| Male | 23 | 16 (70) | 07 (30) | ||
| Female | 5 | 04 (80) | 01 (20) | ||
| Family history | 0.132 | NS | |||
| Absent | 26 | 19 (73) | 07 (27) | ||
| Present | 2 | 01 (50) | 01 (50) | ||
| Anatomic site | -0.045 | NS | |||
| Buccal mucosa | 13 | 09 (69) | 04 (31) | ||
| Tongue | 15 | 11 (73) | 04 (27) | ||
| Tobacco habit | 0.032 | NS | |||
| Absent | 4 | 03 (75) | 01 (25) | ||
| Present | 24 | 17 (71) | 07 (29) | ||
| Notation: NS: Not Significant | |||||
Table 4A: Relationship between SCCAg mRNA expression and patient characteristics (PPBs).
| SCCAg mRNA | |||||
|---|---|---|---|---|---|
| Characteristics | N | <2504.31 N (%) |
>2504.31 N (%) |
r | p value |
| Total patients | 28 | 20 (71) | 08 (29) | ||
| Tumor size | 0.388 | 0.04 | |||
| T1/T2 | 12 | 11 (92) | 01 (08) | ||
| T3/T4 | 16 | 09 (56) | 07 (44) | ||
| Nodal status | 0.024 | NS | |||
| Negative | 18 | 13 (72) | 05 (28) | ||
| Positive | 10 | 07 (70) | 03 (30) | ||
| HG# | 0.068 | NS | |||
| I* | 12 | 09 (75) | 03 (25) | ||
| II** + III*** | 16 | 11 (69) | 05 (31) | ||
| Keratin | 0.122 | NS | |||
| Absent | 1 | 01 (100) | 00 (00) | ||
| Present | 27 | 19 (70) | 08 (30) | ||
| Growthpattern | -0.138 | NS | |||
| Ulcerative | 22 | 15 (68) | 07 (32) | ||
| Exophytic | 6 | 05 (83) | 01 (17) | ||
| LP@ | 0.388 | 0.04 | |||
| Absent | 12 | 11 (92) | 01 (08) | ||
| Present | 16 | 09 (56) | 07 (44) | ||
| VP$ | 0.073 | NS | |||
| Absent | 19 | 14 (74) | 05 (26) | ||
| Present | 9 | 06 (67) | 03 (33) | ||
| Notations: #Histological grade @Lymphatic permeation $Vascular permeation, *Well differentiated **Moderately differentiated ***Poorly differentiated, NS: Not significant | |||||
Table 4B: Relationship between SCCAg mRNA and tumor characteristics (PPBs).
Survival analysis
Univariate and multivariate survival analysis: Univariate and multivariate analysis for RFS and OS was carried out only in total patients expressing SCCAg mRNA in HNLNs and PPBs and these are depicted in Tables 5A, 5B and 7A, 7B, respectively.
| Variables | Wald | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Univariate analysis | |||||
| Tumor size | 14.28 | 5.853 | 2.341 | 14.634 | 0.0001 |
| Histological grade | 0.104 | 1.158 | 0.473 | 2.835 | 0.747 |
| Keratin | 0.968 | 2.747 | 0.367 | 20.563 | 0.325 |
| Growth pattern | 1.416 | 0.295 | 0.039 | 2.204 | 0.234 |
| LP@ | 6.059 | 3.217 | 1.269 | 8.155 | 0.014 |
| VP$ | 0.129 | 1.446 | 0.193 | 10.825 | 0.72 |
| SCCAg mRNA* | 18.274 | 10.273 | 3.531 | 29.83 | 0.0001 |
| Multivariate analysis | |||||
| Step 1 SCCAg mRNA* | 18.274 | 10.273 | 3.531 | 29.83 | 0.0001 |
| Notations: @Lymphatic permeation, $Vascular permeation, *SCCAg mRNA expression in HNLNs. | |||||
Table 5A: Univariate and multivariate survival analysis for RFS-Cox proportional hazard model (HNLNs, N=42).
| Variables | Wald | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Univariate analysis | |||||
| Tumor size | 2 | 2.318 | 0.724 | 7.427 | 0.157 |
| Histological grade | 2.511 | 2.565 | 0.8 | 8.223 | 0.113 |
| Growth pattern | 0.002 | 0.969 | 0.27 | 3.48 | 0.961 |
| LP@ | 3.25 | 3.255 | 0.902 | 1.746 | 0.071 |
| VP$ | 1.129 | 0.288 | 0.615 | 5.144 | 0.288 |
| SCCAg mRNA* | 10.71 | 9.644 | 2.48 | 37.494 | 0.001 |
| Multivariate analysis | |||||
| Step 1 SCCAg mRNA* | 10.71 | 9.644 | 2.48 | 37.494 | 0.001 |
| Notations: @Lymphatic permeation, $Vascular permeation, *SCCAg mRNA expression in PPBs. | |||||
Table 5B: Univariate and multivariate survival analysis for RFS-Cox proportional hazard model (PPBs, N=24).
| N | Patients without relapse |
Patients with relapse | Log rank | p value | |
|---|---|---|---|---|---|
| SCCAg mRNA expression | N (%) | N (%) | |||
| HNLNs | 42 | 22 (52) | 20 (48) | 27.15 | 0.00001 |
| <1901.65 | 28 | 22 (79) | 06 (21) | ||
| >1901.65 | 14 | 00 (00) | 14 (100) | ||
| PPBs | 24 | 10 (42) | 14 (58) | 15.89 | 0.0001 |
| <2504.31 | 16 | 10 (100) | 06 (38) | ||
| >2504.31 | 8 | 00 (00) | 08 (100) |
Table 6: Survival analysis for RFS based on SCCAg mRNA expression in HNLNs and PPBs-Kaplan-Meier survival estimation.
| Variables | Wald | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Univariate analysis | |||||
| Tumor size | 19.912 | 13.712 | 4.342 | 43.305 | 0.0001 |
| Histological grade | 0.154 | 0.811 | 0.284 | 2.312 | 0.695 |
| Growth pattern | 0.393 | 0.522 | 0.068 | 3.989 | 0.531 |
| LP@ | 10.385 | 5.796 | 1.991 | 16.874 | 0.001 |
| VP$ | 0.507 | 2.097 | 0.273 | 16.106 | 0.477 |
| SCCAg mRNA* | 14.306 | 12.303 | 3.351 | 45.171 | 0.0001 |
| Multivariate analysis | |||||
| Step 1 Tumor size | 19.912 | 13.712 | 4.342 | 43.305 | 0.0001 |
| Step 2 SCCAg mRNA* | 5.861 | 6.233 | 1.417 | 27.424 | 0.015 |
| Tumor size | 7.423 | 6.284 | 1.675 | 23.576 | 0.006 |
| Notations: @Lymphatic permeation, $Vascular permeation, *SCCAg mRNA expression in HNLNs. | |||||
Table 7A: Univariate and multivariate survival analysis for OS - Cox proportional hazard model (HNLNs, N=42).
| Variables | Wald | OR | 95% CI | p value | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Univariate analysis | |||||
| Tumor size | 2.179 | 2.411 | 0.749 | 7.754 | 0.14 |
| Histological grade | 3.852 | 3.26 | 1.002 | 10.608 | 0.05 |
| Growth pattern | 0.161 | 1.299 | 0.361 | 4.676 | 0.689 |
| LP@ | 3.613 | 3.482 | 0.962 | 12.605 | 0.057 |
| VP$ | 1.586 | 1.982 | 0.684 | 5.747 | 0.208 |
| SCCAg mRNA* | 7.254 | 4.465 | 1.503 | 13.267 | 0.007 |
| Multivariate analysis | |||||
| Step 1 SCCAg mRNA* | 7.254 | 4.465 | 1.503 | 13.267 | 0.007 |
| Notations: @Lymphatic permeation, $Vascular permeation, *SCCAg mRNA expression in PPBs. | |||||
Table 7B: Univariate and multivariate survival analysis for OS-Cox proportional hazard model (PPBs, N=24).
Relapse-free survival
Histological negative lymph nodes (HNLNs): In total patients, univariate analysis of the significant survival data obtained from HNLNs when analyzed along with histopathological parameters of primary tumors showed that the significant risk predictors were tumour size (OR=5.85, p=0.0001), lymphatic permeation (OR=3.24, p=0.014) and SCCAg mRNA (OR=10.27, p=0.0001). However, in multivariate analysis, only SCCAg mRNA expression in HNLNs was a significant predictor of RFS (OR=10.27, p=0.0001). The relative risk of recurrence due to oral cancer was 10.27 in patients with SCCAg mRNA expression >1901.65 as compared to those with SCCAg mRNA expression <1901.65 (Table 5A).
Pretherapeutic peripheral blood samples (PPBs): Keratin was not included in analysis because all patients showed presence of keratin. In both, univariate and multivariate analysis for RFS, only SCCAg mRNA expression in PPBs was the most significant prognostic factor (OR=9.64, p=0.001). The relative risk for recurrence due to oral cancer was 9.64 times more in patients with SCCAg mRNA expression >2504.31 as compared to those with SCCAg mRNA expression <1901.65 (Table 5B).
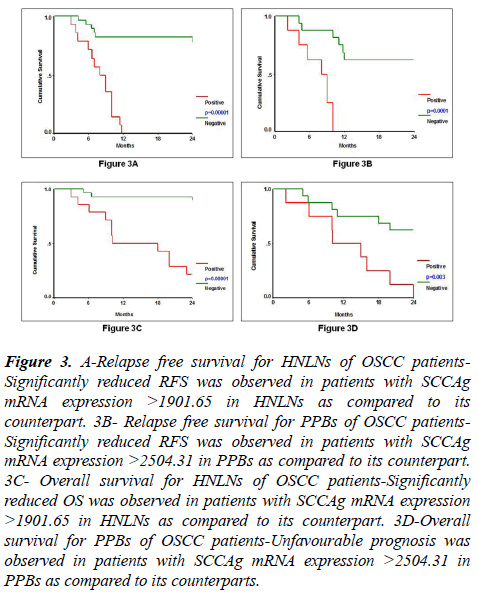
Relapse-free survival was evaluated by Kaplan-Meier survival estimation in HNLNs (N=42) and PPBs (N=24) based on their cut-off value for SCCAg mRNA expression which was 1901.65 and 2504.31, respectively. SCCAg mRNA expression was a significant prognostic factor for RFS in HNLNs (Log rank=27.15, df=1, p=0.00001) and in PPBs (Log rank=15.89, df=1, p=0.0001) (Table 6, Figures 3A and 3B). Hundred percent (14/14) of patients with SCCAg mRNA expression >1901.65 in their HNLNs showed significantly reduced RFS as compared to 21% (6/28) of patients with SCCAg mRNA expression <1901.65. Similarly, 100% (8/8) of patients with SCCAg mRNA expression >2504.31 in PPBs showed significantly reduced RFS as compared to 38% (6/16) of patients with SCCAg mRNA expression <2504.31.
Figure 3: A-Relapse free survival for HNLNs of OSCC patients- Significantly reduced RFS was observed in patients with SCCAg mRNA expression >1901.65 in HNLNs as compared to its counterpart. 3B- Relapse free survival for PPBs of OSCC patients- Significantly reduced RFS was observed in patients with SCCAg mRNA expression >2504.31 in PPBs as compared to its counterpart. 3C- Overall survival for HNLNs of OSCC patients-Significantly reduced OS was observed in patients with SCCAg mRNA expression >1901.65 in HNLNs as compared to its counterpart. 3D-Overall survival for PPBs of OSCC patients-Unfavourable prognosis was observed in patients with SCCAg mRNA expression >2504.31 in PPBs as compared to its counterparts.
Overall survival
Histologically negative lymph nodes (HNLNs): A survey of histopathological variables and SCCAg mRNA expression in HNLNs in relation to OS, demonstrated that factors affecting OS were tumor size (OR=13.71, p=0.0001), lymphatic permeation (OR=5.79, p=0.001) and SCCAg mRNA (OR=12.30, p=0.0001). However, multivariate survival analysis revealed that tumor size (OR=13.71, p=0.0001) at step 1 and SCCAg mRNA in the HNLNs (OR=6.23, p=0.015) at step 2 contributed to OS (Table 7A).
Pretherapeutic peripheral blood samples (PPBs): In univariate analysis, the prognosticators that were associated with OS were histologic grade (OR=3.26, p=0.05) and SCCAg mRNA expression in PPBs (OR=4.46, p=0.007). In multivariate analysis, the expression of SCCAg mRNA in PPBs emerged as the most significant factor for predicting OS (OR=4.46, p=0.007). The relative risk for death due to oral cancer was 4.46 times more in patients with SCCAg mRNA expression >2504.31 as compared to those with SCCAg mRNA expression <2504.31 (Table 7B).
Kaplan-Meier survival estimation for OS was studied in HNLNs (N=42) and in PPBs (N=24) according to the cut-off value of SCCAg mRNA in HNLNs (1901.65) and PPBs (2504.31). High level of SCCAg mRNA expression in HNLNs (Log rank=22.60, p=0.00001) and PPBs (Log rank=8.69, p=0.003) was associated with inferior OS (Table 8, Figures 3C and 3D). Seventy-nine percent (11/14) of patients with SCCAg mRNA expression >1901.65 in HNLNs showed significantly shorter OS as compared to 11% (3/28) of patients with SCCAg mRNA expression <1901.65. Similarly, 100% (8/8) of patients with SCCAg mRNA expression >2504.31 in PPBs showed significantly poorer OS as compared to 38% (6/16) of patients with SCCAg mRNA expression <2504.31.
| Patients alive | Patients died | Log rank | p value | ||
|---|---|---|---|---|---|
| SCCAg mRNA expression | N | N (%) | N (%) | ||
| HNLNs | 42 | 28 (67) | 14 (33) | 22.6 | 0.00001 |
| <1901.65 | 28 | 25 (89) | 03 (11) | ||
| >1901.65 | 14 | 03 (21) | 11 (79) | ||
| PPBs | 24 | 10 (42) | 14 (58) | 8.69 | 0.003 |
| <2504.31 | 16 | 10 (63) | 06 (38) | ||
| >2504.31 | 8 | 00 (00) | 08 (100) |
Table 8: Survival analysis for overall survival based on SCCAg mRNA expression in HNLNs and PPBs-Kaplan-Meier survival estimation.
Discussion
In the current study, we aimed at determining the clinical relevance of SCCAg mRNA as a marker for detecting micrometastasis. The presence of metastatic squamous cells in the cervical LNs of patients with OSCC reflects disease progression and implies that the disease is no longer localized. Further, in these patients the chance of long-term control is decreased by 50% when compared to patients who have similar primary tumors without nodal metastasis [21]. Hence, in OSCC, the status of cervical LNs is an extremely important prognostic factor used in the clinical management of the disease. Routine histological examination may fail to detect low number of tumor cells, which is however, possible by molecular diagnosis. Given this, early detection of micrometastasis in node negative OSCC patients is expected to assist in treatment planning and thereby improve survival. Several studies have revealed presence of micrometastatic tumor cells in the LNs after staining with epithelial specific antibodies [10-13] and molecular analysis [17-19,22,23] in head and neck cancers. However, using molecular analysis, minimal data exists on the prognostic relevance of micrometastasis in HNLNs or PPBs of OSCC or their association with clinicopathological features of the primary tumor [24,25] but there are no reports that have evaluated the prognostic value of SCCAg mRNA or correlated the expression of SCCAg mRNA with clinicopathological parameters in OSCC. Our data provided important insight into the possible clinical application of micrometastasis using SCCAg mRNA expressed exclusively by squamous cell tissues.
In the present study, the nested RT-PCR results showed absence of SCCAg mRNA expression in the LNs and peripheral blood samples of the control group, indicating a very high rate of specificity. Similar specificity for SCCAg mRNA expression was observed by Hamakawa et al. in head and neck cancer and by Kano et al. and Kaganoi et al. in esophageal cancer. Concerning sensitivity, various investigating groups have evaluated the detection threshold of SCCAg mRNA in LNs and PPBs [13,19,26,27]. Our data demonstrated that SCCAg mRNA could detect micrometastasis in 27% and 29% of HNLNs and PPBs, respectively, using the sensitive nested RT-PCR assay and therefore may serve as a useful molecular marker for detecting micrometastasis. Studies demonstrating the efficacy of molecular markers to detect micrometastasis in HNLNs and PPBs in various malignancies have already been reported [28]. In HNSCC, using conventional RT-PCR, Hamakawa et al. and Onishi et al. reported SCCAg mRNA in 18.7% and 7.4% of HNLNs, respectively. With quantitative RT-PCR, 17.6% HNLNs expressed SCCAg mRNA [16]. Based on cytokeratin 5 and 13 gene expression, McDonald et al. and Hamakawa et al. were able to detect micrometastasis in 40% and 14.4% of HNLNs, respectively. Compared to these studies examining several LNs from a single patient, our study examined only one LN per individual patient. These results emphasize the importance of verifying the HNLNs for micrometastasis. With regard to detection of circulating tumor cells, there are very few reports in HNSCC that have examined the importance of molecular markers for detecting micrometastatic cells in circulation. In head and neck cancer, Brakenhoff et al. applying E48 was able to detect micrometastasis in 10% of the blood samples. In OSCC, Zen et al. showed the presence of SCCAg mRNA and EGFR mRNA in the circulation using both conventional and quantitative RT-PCR. In other malignancies, SCCAg mRNA detected circulating tumor cells in 40% and 33% of cervical and esophageal cancer, respectively [27,29]. Thus, the detection of SCCAg mRNA positive cells in PPBs in oral cancer patients is an indication of dissemination of the tumor cells. Together, these results are in agreement with the clinical data indicating a 50% decrease in survival in node negative HNSCC patients (Snow et al., Zatterstrom et al., Leemans et al.) due to loco-regional recurrence or distant metastasis, further suggesting that these patients have micrometastasis and were under staged. The under staging is a well known phenomenon as demonstrated by serial sectioning of routine HNLNs and by immunohistochemistry for different epithelial molecules as well as the molecular techniques for detection of mRNA of specific molecular markers [11-13,26,30].
SCCAg is identified to be a member of ova-albumin family of serine protease inhibitor [31] and shares high amino acid sequence homology with plasminogen activator type 2 [32]. Since plasminogen activators have been reported to play a crucial role in malignant behavior [33-35], it is quite likely that SCCAg may take some part in the invasion or metastasis of tumor cells. Hence, it would be interesting to note if SCCAg showed any association with clinicopathological features of the primary tumor. In this series, SCCAg mRNA expression in HNLNs and PPBs when correlated with clinicopathological parameters of the primary tumor, showed a significant positive correlation with tumor size and lymphatic invasion, suggesting that SCCAg mRNA may be associated with progression of primary oral tumors that have a more aggressive clinical course. Further, although, HNSCC is thought to metastasize initially via the lymph as judged from clinical disease progression, we observed no correlation between SCCAg mRNA analyzed in the HNLNs and PPBs and even when HNLNs and PPBs were assessed from the same set of node negative OSCC patients (N=15). One of the probable reasons could be the existence of several different pathways and anastomosis of lymphatics in the neck (van den Brekel et al.), and in this study, only one LN per patient was analyzed. To our knowledge there are no reports that have correlated SCCAg mRNA with clinicopathological prognosticators in oral cancer. However, these results are consistent with the findings of Kaganoi et al. demonstrating a correlation with tumor size and venous invasion in esophageal cancer, further confirming the association of SCCAg mRNA with the biological behavior of the tumor.
Since node negative patients who undergo successful radical resection often encounter recurrence or distant metastasis, it is important to assess whether genetic diagnosis for micrometastasis is associated with prognosis. Multivariate statistical analysis of RFS in relation to SCCAg mRNA in HNLNs and PPBs revealed a relative risk of 10.27 and 9.64, respectively, for developing metastasis in patients with SCCAg mRNA expression compared to those without SCCAg mRNA expression. Moreover, SCCAg mRNA was also able to predict OS. In line with this, Kaganoi et al. reported SCCAg mRNA to be a significant predictor of prognosis in esophageal cancer. Even, in various carcinomas, it has been demonstrated that detection of micrometastasis has a significant impact on patient survival [36-39]. In HNSCC, Nieuwenhuis et al. in pathological N0 group demonstrated that the presence of E48 positive LNs was significantly associated with a distinctly poor cause specific survival as compared to those with E48 negative LNs. Also, recently, Williams et al. have demonstrated that elevated levels of SCCAg were associated with a reduced chance of achieving a complete response and an increased chance of recurrence and death in anal canal patients. Hence, SCCAg mRNA may also be regarded as a clinically relevant prognostic factor for early detection of micrometastasis in OSCC.
In conclusion, SCCAg mRNA analyzed by nested RT-PCR may serve as an important marker for assessment of micrometastasis that contributes to poor prognosis in node negative OSCC patients. Further, it may represent a useful tool for more accurate staging, which could improve disease management and help to obtain maximal therapeutic benefit from adjuvant therapies.
Acknowledgements
Authors are thankful to the Gujarat Cancer Society and The Gujarat Cancer Research Institute for providing financial support for the fulfillment of this study.
References
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-86.
- Wang Q, Gao P, Wang X, et al. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. Sci Rep. 2014;30(4):6802.
- Petersen PE. Oral cancer prevention and control–the approach of the World Health Organization. Oral oncol. 2009;45(4-5):454-60.
- Sankara NR, Ramadas K, Thomas G, et al. Effect of screening on oral cancer mortality in Kerala, India: A cluster randomized controlled trial. Lancet. 2005;365(9475):1927-33.
- Argiris A, Karamouzis MV, Raben D, et al. Head and neck cance