Research Article - Journal of RNA and Genomics (2024) Volume 20, Issue 1
Unveiling the role of lncRNA Nron and Nfatc3 in Type 2 Diabetes Mellitus: A molecular insight for disease pathogenesis and biomarker discovery.
Zivar Salehi1*, Diba Zebardast1, Mona Zaersabet1, Kiana Sojoudi1, Farhad Mashayekhi1, Behrang Motamed2
1Department of Biology, University of Guilan, Rasht, Iran
2Department of Internal Medicine, Guilan University of Medical Sciences, Rasht, Iran
- Corresponding Author:
- Zivar Salehi
Department of Biology, University of Guilan, Rasht, Iran
E-mail: salehiz@guilan.ac.ir
Received: 20-Jan-2024, Manuscript No. RNAI-24-125550; Editor assigned: 23-Jan-2024, Pre QC No. RNAI-24-125550(PQ); Reviewed: 07-Feb-2024, QC No. RNAI-24-125550; Revised: 14-Feb-2024, Manuscript No. RNAI-24-125550(R); Published: 21-Feb-2024, DOI: 10.35841/2591-7781.19.1000176.
Citation: Zivar Salehi. Unveiling the role of lncRNA Nron and NFATc3 in Type 2 Diabetes Mellitus: A molecular insight for disease pathogenesis and biomarker discovery. J RNA Genomics 2024;20(1):1-9.
Abstract
Long non-coding RNAs (lncRNAs) are key factors in the progression of many metabolic diseases. This study aimed to investigate the role of long non-coding RNA repressor of the nuclear factor of activated T cells (lncRNA NRON) and Nuclear Factor of Activated T-Cells, Cytoplasmic 3 (NFATc3) in Type-2 Diabetes Mellitus (T2DM). The expression patterns of NRON and NFATc3 were measured using quantitative real-time PCR and western blotting. Network analysis and gene enrichment analysis were carried out using bioinformatic tools and the Cytoscape platform. The results showed that NRON was upregulated in T2DM patients, while NFATc3 gene and protein accounted for a lower expression (p<0.05). Bioinformatics analysis displayed that these biomarkers play key roles in multiple mechanisms and pathways linked to the development of T2DM. lncRNA NRON and NFATc3 can act as potential biomarkers for the diagnosis and prognosis of T2DM.
Keywords
Diabetes mellitus; Long non-coding RNAs; LncRNA NRON; NFATc3; Gene expression
Introduction
Type 2 Diabetes Mellitus (T2DM), a prevalent metabolic disorders in the population, is defined by elevated levels of blood glucose, which is mainly triggered by a failure in insulin secretion, along with impaired response to insulin by insulinsensitive tissues [1]. In recent decades, the incidence of adult diabetes and glucose intolerance has been rising globally [2]. Studies have postulated that there are over 800,000 new cases of T2D in the Iranian population per year, and by 2030, it is predicted that the number of Iranians with T2D will be approximately 9.2 million [3]. A combination of various genes and environmental factors can trigger this disease [4]. The principal environmental elements of T2DM prevalence are obesity, low physical activity, and a high-calorie diet [1]. Moreover, many studies have identified various functional and positional candidate genes involved in T2DM, with only a few exhibiting a significant and reproducible association [5]. Research showed that the improper expression of long noncoding RNAs (lncRNAs) has a crucial role in controlling pancreatic ß-cell function leading to T2D. LncRNAs, a form of non-coding RNA with over 200 nucleotides, contribute to various biological mechanisms, such as gene expression regulation in different stages, ranging from transcription and translation to RNA degradation and epigenetic changes, as evident in both nucleus and cytoplasm [6-8]. Emerging evidence has suggested that some lncRNAs are vital for the pathophysiology of many cardiovascular disorders, including diabetic cardiomyopathy [9].
Several studies have demonstrated the effects of dysregulated lncRNAs in diabetes mellitus and their involvement in the pathogenesis of this disease so far. For instance, it has been suggested that dysregulated lncRNAs, by regulating inflammation, insulin resistance and secretion, could be involved in T2D pathogenesis [10]. A meta-analysis displayed that the aberrant expression of lncRNA is significantly linked to T2DM pathogenesis [11]. However, there is still scant information on RNA-mediated regulatory networks in T2DM [12]. The lncRNA Non-coding Repressor of NFAT (NRON), mediating various pathophysiological mechanisms, is reported to serve as a cytoplasmic trap for phosphorylated Nuclear Factor of Activated T cells (NFAT) proteins in T cells [13,14]. Furthermore, NRON has been shown to modulate NFAT signaling in different biological processes and diseases, and to suppress it by inhibiting NFAT nucleocytoplasmic shuttling [15]. NFAT, known as a transcription factor with different regulatory roles, like cell survival, differentiation, invasion, angiogenesis, and migration, is involved in many intracellular and extracellular signaling pathways. It is also vital to the incidence of atherosclerosis in diabetic patients and can offer a novel therapeutic target for vascular disorders. The NFAT family has five members: NFATc1, NFATc2, NFATc3, NFATc4, and NFAT5, which has been shown to be expressed in ß-cells.
In the islet, ß-cell function-related genes, especially those involved in glucose sensitivity and biogenesis of insulin secretory granules, are regulated by NFAT. Among NFATc isoforms, NFATc3 is most abundantly found in mouse and human islets. Studies has exhibited that calcineurin/NFAT signaling regulates ß-cells proliferation and secretion [16-20].
Materials and Methods
Study subjects
The subjects consisted of 250 participants recruited from Razi Hospital of Rasht, which is affiliated to Guilan University of Medical Sciences in north of Iran, between March 2017 and September 2019. Among these subjects, 100 were diagnosed with T2DM, and 150 healthy subjects were considered as non- T2DM controls. The T2DM diagnosis was based on criteria set by the World Health Organization (WHO) including Fasting Plasma Glucose (FPG) ≥ 126 mg/dL and/or 2-h Postprandial Glucose (2hPG) ≥ 200 mg/dL. The exclusion criteria were chronic diseases, pregnancy, cancer, inflammatory diseases, Type 1 Diabetes Mellitus (T1DM), and smoking. All participants were long-term residents of Rasht, Iran, with no blood relations. This research was approved by ethics committee of Guilan University of Medical Sciences, Rasht, Iran, and conducted in accordance with the ethical standards stipulated in the most recent revision of the Declaration of Helsinki. All participants provided informed consent before the study commenced.
Clinical information and biochemical measurements
For all subjects, peripheral blood (3 ml) was withdrawn following an overnight fast and gathered in EDTA-containing tubes. The serum was assembled by centrifuging blood samples at 2000 rpm and stored at −80°C for further process. Body Mass Index (BMI), defined as weight (kg) divided by the square of height (m), was also measured. The Glycosylated Hemoglobin A1c (HbA1C), High-Density Lipoprotein Cholesterol (HDL-C), Fasting Plasma Glucose (FPG), Low- Density Lipoprotein Cholesterol (LDL-C), Total Cholesterol (TC) and Triglyceride (TG) were determined by the Laboratory Department of Razi Hospital.
Total RNA extraction
The stored serum samples of subjects in the case and control groups were thawed. For the extraction of total RNA from white blood cells, TRIzol reagent (Invitrogen, USA) was utilized consistent with the manufacturer’s protocols. To analyse the purity and concentration of RNA by the A260/280 ratio, a NanoDrop instrument (Thermos Scientific, USA) was used. The extracted RNA was stored at -70°C in RNase-free tubes for preparation of cDNA.
Quantitative real-time PCR (qRT-PCR) analysis and cDNA synthesis
cDNA was generated from the RNA derived from T2DM patients and healthy controls using cDNA Synthesis Kit (Thermo Scientific, USA). Following the reverse transcription reaction, Real-time PCR was applied using SYBR green to the prepared cDNA to amplify the target fragment and estimate the gene expression quantitatively. The synthesis of primers for LncRNA NRON, NFATc3, and the housekeeping gene Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) conducted by Metabion Co. (Bavaria, Germany), the sequences of which are presented in Table 1.
| Gene | Sequence (5′ to 3′) | Number of bases (bp) |
|---|---|---|
| LncRNA NRON | Sense: 5ʹ-GTTGCTTTACTGAGATGACACAGG-3ʹ | 24 |
| Antisense: 5ʹ-GAGGAAAACCCATTCTACAGTCAG-3ʹ | 24 | |
| NFATC3 | Sense: 5ʹ-CCACACCCCTATATTTCGCAC-3ʹ | 21 |
| Antisense: 5ʹ-CAGGAGCTTCACAACAGGATG-3ʹ | 21 | |
| GAPDH | Sense: 5ʹ-CATCACCATCTTCCAGGAGCG-3ʹ | 21 |
| Antisense: 5ʹ-GGAGGCATTGCTGATGATCTTG-3ʹ | 22 |
Table 1: Primers used for quantitative Real-time PCR (qRT-PCR).
Real-time PCR was run on a Roche lightcycler 96 Instrument (Roche Molecular Systems) using 20 μl reaction volume for 45 cycles, including initial 20 s denaturation at 95°C, 30 s annealing at 60°C, and 10 s extension at 72°C . To terminate the reaction, a final ending step was added at 72°C for 5 min. In the negative control group, water was utilized instead of cDNA and each reaction was performed in triplicate. The relative gene expression was evaluated by the ΔΔCT method, and normalization to GAPDH as an endogenous control gene.
Western blotting
For the isolation of Peripheral Blood Mononuclear Cells (PBMCs), the Ficoll isolation method was used [21]. PBMCs were lysed using a buffer of Radio immunoprecipitation Assay (RIPA) (R0278, Sigma-Aldrich) containing a mixture of protease inhibitors. The centrifuge of homogenate was carried out at 12,000 g for 20 min at 4°C, and the resulting supernatant was gathered. Following quantification with the Pierce BCA Protein Assay Kit (Thermo Scientific), the analysis of lysates containing 40 μg of proteins was performed by 10% SDS-PAGE and proteins were transferred onto a polyvinylidene difluoride membrane (Bio-Rad) using a semi-dry system (Biorad) and the transfer solution (Tris 48 mM, SDS 0.0375%, glycine 39 mM, and methanol 20%). We then blocked the blots in 5% non-fat milk that was diluted in Tris-Buffered Saline (TBS) comprising 0.1% (v/v) Tween-20 (TBS-T) and investigated overnight by means of antibodies against primary antibodies, such as NFATc3 antibody (1:1000 dilution; 18222-1-AP,Proteintech) and β-actin (1:1000 diluted; A5316, Sigma) at 4˚C. After washing membranes with TBS-T buffer three times, they were incubated in a blocking buffer with secondary antibodies coupled to horseradish peroxidase (1:5000, Amersham Biosciences, USA) for 2 h at room temperature. For the quantification of protein bands, ImageJ 1.46r software (NIH, Bethesda, MD) was used, and the mean densities of pixels in the protein bands were measured. Relative light densities of the protein bands were computed and stated as a ratio to ß-actin.
Statistical analysis
GraphPad Prism software version 8.0 (GraphPad Software, Inc.) and SPSS software (version 24, SPSS Inc, Chicago, IL) were used for statistical analysis. The independent t-test was utilized to measure inter-group significance (p<0.05). Values are expressed as mean ± standard deviation for n=3. All graphical representations were also prepared using GraphPad Prism. The association of lncRNA NRON expression with NFATc3 and clinical indicators was assessed using Pearson correlation analysis.
In Silico analyses
Identification of lncRNA NRON target genes involved in type 2 diabetes mellitus
The database of National Center for Biotechnology Information (NCBI) was used to find proteins associated with type 2 diabetes mellitus in Homo sapiens. LncRNA NRON target genes were predicted using NPInter, RNAInter, LncRNADisease, and RAID databases. Then, the predicted targets for NRON were compared with the proteins related to type 2 diabetes mellitus, and Cytoscape software (version 3.9.1) was used to visualize their regulatory network.
Construction of the Protein-Protein Interaction (PPI) network
NRON target genes were subjected to the Search Tool for Retrieval of Interacting Genes (STRING) in Cytoscape Software (version 3.9.1) in an attempt to find their PPI network. For this purpose, STRING: Protein query was used as data source to create a network between 67 proteins targeted by NRON. Informed by the molecular action and nodes, the edge score was computed with confidence score over 0.400. The Cytoscape platform was used for the creation, design, and organization of network.
ClueGO/CluePedia Enrichment and PANTHER Analysis
The Cytoscape plugin ClueGO/CluePedia (v.2.5.9) was used to study the enriched pathways linked to the Kyoto Encyclopaedia of Genes and Genome (KEGG) database. The statistical options for ClueGO enrichment analysis were set based on an enrichment/depletion method with a two-sided hypergeometric test with p ≥ 0.05, corrected with the Bonferroni step down for each p-value calculation, and kappa score ≥ 0.4 was considered as a primary criterion. Enriched pathways with a p-value ≤ 0.05 were deemed significant. Furthermore, PANTHER database) was utilized for gene ontology analysis of NFATc3.
Results
Demographic and metabolic characteristics of subjects
Table 2 represents major clinical features of T2DM subject in the intervention and control groups. The present study included 100 patients with T2DM (age range of 40-65 years; 48% males) and 150 non-T2DM controls (age range of 38-66 years; 41% males). T2DM subjects had substantially increased levels of FPG and HbA1c in comparison to the healthy group (p<0.0001). In contrast, no statistically significant difference was observed between HDL-C, LDL-C, TG, and TC concentrations.
| Parameters | Control (n=150) | T2DM (n=100) | P-value |
|---|---|---|---|
| Age (year) | 52.0 ± 7.1 | 187.5 ± 22.1 | 0.091 |
| Gender (male/female) | 61/89 | 48/52 | 0.579 |
| BMI (kg/m2) | 24.9 ± 3.3 | 25.8 ± 2.8 | 0.284 |
| HbA1c (%) | 4.9 ± 0.5 | 9.4 ± 1.7 | <0.0001 |
| FPG | 95.2 ± 18.1 | 210.1 ± 76.0 | <0.0001 |
| HDL-C (mg/dL) | 47.6 ± 8.2 | 45.3 ± 8.0 | 0.294 |
| LDL-C (mg/dL) | 139.3 ± 16.3 | 145.7 ± 25.5 | 0.266 |
| TG (mg/dL) | 139.3 ± 16.3 | 175.4 ± 26.1 | 0.378 |
| TC (mg/dL) | 187.5 ± 22.1 | 197.8 ± 25.1 | 0.113 |
Note: Data are expressed as mean ± SD, and were analysed by an independent samples t test; P<0.05 was considered statistically significant. BMI, body mass index; HbA1c, hemoglobin A1c; FPG, fasting plasma glucose; HDL-C, high-density lipoprotein cholesterol; LDL-C, low-density lipoprotein cholesterol; TG, triglyceride; TC, total cholesterol
Table 2: General characteristics of enrolled patients with T2DM and control group
Expression profiles of lncRNA NRON and NFATc3 in T2DM patients and controls
To analyse the expression profiling of lncRNA NRON and NFATc3 gene in patients with T2DM and healthy controls, Real-time PCR was used Figures 1a and 1b. The findings exhibited a significantly (p<0.0001) higher expression level of lncRNA NRON in T2DM cases than in the healthy controls (1.87 ± 0.035 and 1.01 ± 0.028, respectively). In comparison, the evaluation of the NFATc3 transcript level reflected a substantial discrepancy in the gene expression patterns of this marker between the two groups (p<0.0001). T2DM patients displayed a statistically decreased level of NFATc3 expression (1.00 ± 0.062) compared to the healthy group (2.10 ± 0.09).
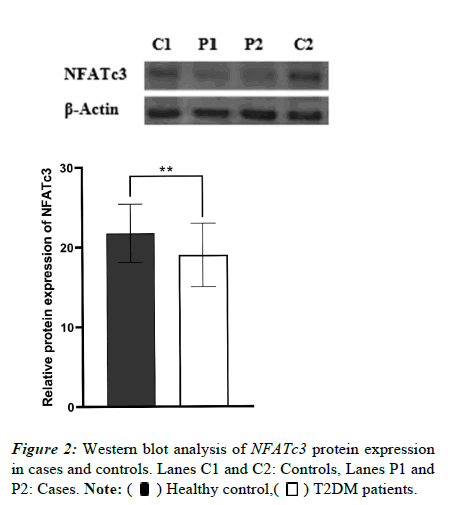
We also investigated NFATc3 protein expression using western blotting. As shown in Figure 2, NFATc3 expression in blood lymphocytes dropped in diabetic cases as opposed to the controls (p=0.0017).
Correlation of lncRNA NRON with NFATc3 and clinical indicators in T2DM patients
The correlations between lncRNA NRON, NFATc3 gene, and clinical characteristics were assessed in the cohort of patients. The findings suggested that lncRNA NRON was negatively associated with NFATc3 expression (p=0.001). In addition, there was a positive correlation between HbA1c and FPG and lncRNA NRON and a negative correlation with NFATc3 gene expression was also observed (p<0.05). More detail in terms of correlation tests is listed in Table 3.
| Variables | lncRNA NRON | NFATc3 | ||
|---|---|---|---|---|
| r | p | r | p | |
| NFATc3 | -0.214 | 0.001 | -- | -- |
| Age (year) | -0.045 | 0.82 | 0.367 | 0.08 |
| BMI (kg/m2) | 0.276 | 0.155 | 0.223 | 0.253 |
| HbA1c (%) | 0.002 | 0.155 | -0.124 | 0.026 |
| FPG | 0.36 | 0.009 | -0.548 | 0.013 |
| HDL-C (mg/dL) | 0.176 | 0.371 | -0.192 | 0.326 |
| LDL-C (mg/dL) | 0.024 | 0.902 | 0.203 | 0.3 |
| TG (mg/dL) | -0.174 | 0.375 | -0.009 | 0.965 |
| TC (mg/dL) | -0.177 | 0.367 | -0.058 | 0.768 |
BMI: Body Mass Index; HbA1c: Hemoglobin A1c; FPG: Fasting Plasma Glucose; HDL-C: High-Density Lipoprotein Cholesterol; LDL-C: Low-Density Lipoprotein Cholesterol; Tg: Triglyceride, Tc: Total Cholesterol.
Table 3: Correlations of lncRNA NRON and NFATc3 expression with each other and biochemical parameters.
PPI Network construction
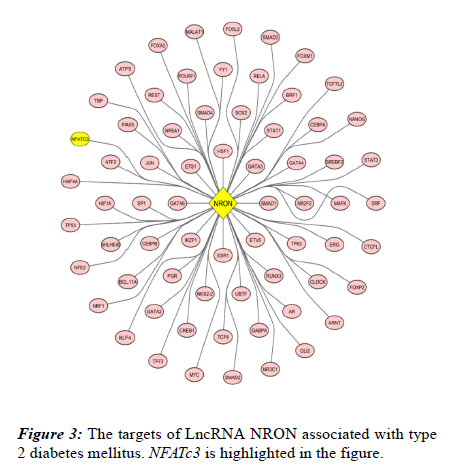
Through bioinformatics analysis, 67 biomarkers involved in T2DM were identified and directly targeted by NRON (Figure 3).
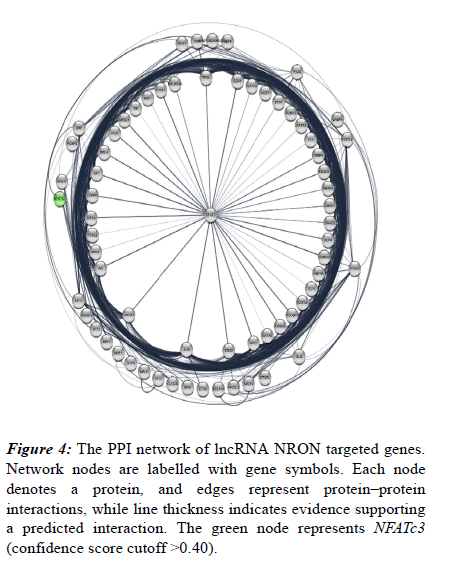
Also, their protein-protein interaction was analysed using STRING database following visualization performed by Cytoscape software. Accordingly, a total of 683 edges were predicted for the 67 targets (Figure 4). Network nodes are identical to proteins and edges correspond to the proteinprotein interactions (post-translational modifications and splice isoforms are collapsed, namely, each node denotes the entire proteins created by a single, protein-coding gene locus). Proteins account for a shared function and relations are intended to be both specific and meaningful. Based on the results derived from the network, NFATc3 is also associated with this process and include multiple interactions with other involved proteins. NFATc3 is shown as a green node in the network (Figure 4).
Figure 4: The PPI network of lncRNA NRON targeted genes. Network nodes are labelled with gene symbols. Each node denotes a protein, and edges represent protein–protein interactions, while line thickness indicates evidence supporting a predicted interaction. The green node represents NFATc3 (confidence score cutoff >0.40).
Functional Enrichment Analysis
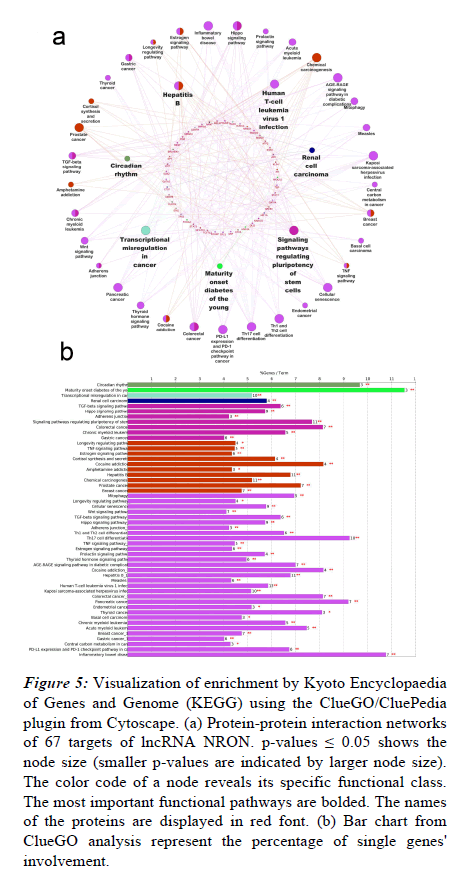
To investigate the enriched pathways linked to the NRON targets, the list of identified biomarkers was imported into the ClueGO app of the Cytoscape platform for the functional enrichment analysis. As shown in Figure 5a, multiple functions were enriched for the targeted proteins, the most significant of which are signaling pathways, maturity-onset diabetes of the young, regulating stem cells’ pluripotency, renal cell carcinoma, circadian rhythm, hepatitis B, transcriptional misregulation in cancer, and human T-cell leukemia virus-1 infection. Figure 5b shows the percentages of significantly overrepresented KEGG terms.
Figure 5: Visualization of enrichment by Kyoto Encyclopaedia of Genes and Genome (KEGG) using the ClueGO/CluePedia plugin from Cytoscape. (a) Protein-protein interaction networks of 67 targets of lncRNA NRON. p-values ≤ 0.05 shows the node size (smaller p-values are indicated by larger node size). The color code of a node reveals its specific functional class. The most important functional pathways are bolded. The names of the proteins are displayed in red font. (b) Bar chart from ClueGO analysis represent the percentage of single genes' involvement.
In addition, the NFATc3 was separately analysed for functional classification using the PANTHER tools (Protein Analysis Through Evolutionary Relationships, Foster City, CA, USA). Accordingly, NFATc3 is involved in a verity of pathways, such as axon guidance mediated by netrin (16.7%), B-cell activation (16.7%), T-cell activation (16.7%), gonadotropin-releasing hormone receptor pathway (16.7%), inflammation mediated by chemokine and cytokine signaling pathway (16.7%), and Wnt signaling pathway (16.7%). Also, it plays a role in biological regulation (16.7%), cellular process (16.7%), metabolic process (16.7%), multicellular organismal process (16.7%), response to stimulus (16.7%), and signaling (16.7%).
Discussion
LncRNAs appear to be a likely biomarker for the prediction and diagnosis of disorders, because some complicated diseases, such as diabetes mellitus, demonstrate a dysregulated expression. It has been revealed that type 2 diabetes is 12 times more likely in people with a lower expression of lncRNA GAS5 [22,23]. Moreover, the relative expression of lncRNA KCNQ1OT1 was significantly higher in the T2D group than in the control group [24]. also reported an elevated expression profile of lncRNA NR2F1-AS1 in uncomplicated T2D patients [25]. On the other hand, glucose homeostasis maintained by pancreatic ß-cells through insulin secretion indicates the pivotal role of ß-cells. Research shows that NFATs can regulate the development and proliferation of ß-cells in human and mouse islets. Moreover, among all isoforms of NFATc, NFATc3 has the highest expression in islets, which reflects the underlying role of this marker in ß-cells function and blood sugar balance [26].
In the current study, lncRNA NRON and its target genes, including NFATc3 were selected to evaluate their association with the development of T2DM. As far as we are concerned, there is a paucity of research on expression regulation of lncRNA NRON and NFATc3 in Type 2 diabetes mellitus. Herein, the mRNA transcript level of lncRNA NRON was higher in the patient group than in the healthy group. In contrast, qRT-PCR and Western blotting displayed a decreased expression of NFATc3 gene and protein in T2DM. Hu et al., induced diabetes in mice fed with a high-fat diet and then examined changes in NFATc3 expression. The results revealed that NFATc3 expression levels were downregulated in pancreatic β-cells of HFD-fed mice, which may be associated with insulin secretion impairments and diabetes [26]. Li studied the association between lncRNA NRON and NFATc3 in atrial fibrosis. They found that the overexpression of lncRNA NRON, by inhibiting NFATc3 nuclear transport, can suppress the expression of miR-23a, leading to the alleviation of myocardial fibrosis [27]. Previous research demonstrated that NRON, by blocking the nucleation of NFATc3, inhibited Interleukin (IL)-12 expression and transcription in myocardial cells, which contributes to the weakening of atrial fibrosis [28]. Furthermore, the results of correlation found in our study showed that lncRNA NRON upregulation was associated with NFATc3 downregulation, and positively correlated with HbA1c, FPG, HDL-C. Wang et al., implicated a correlation between three upregulated lncRNAs and metabolic variables, including FPG, HbA1c, BMI, 2hPG, and HOMA-IR [29]. Jiang et al., reported an association between two candidate lncRNAs (lnc- HIST1H2AG-6 and lnc-AIM1-3) and the levels of FIN, FPG, HOMA-B and HbA1c-B [30]. In addition, a negatively correlation was observed between NFATc3 expression and HbA1c, FPG. Another study has demonstrated the significant association of NFATc1 with metabolic indices [31].
We used bioinformatic tools to further validate the present study in terms of screening the target genes of lncRNA NRON involved in T2DM development. One of the NRON targeted genes predicted to be associated with this process, was NFATc3. The results of bioinformatic analysis showed that the target genes of lncRNA NRON can play a significant role in T2DM incidence and development through different pathways and mechanisms (Figure 5a). One of the noticeable pathways is the Wnt signaling pathway, which is vital for the activation of JNK, TCF7L2 variants, and gene expression caused by β- catenin/TCF7L2, as well as calcineurin/NFAT. As such, it is significantly correlated with T2DM and related syndromes [32]. Other roles identified for NRON target genes are Th2 and Th1 cell differentiation. The balance between Th1 and Th2 cell differentiation is important for the development and progression of type 2 diabetes, and while an imbalance in favor of Th1 cells promotes inflammation and insulin resistance, an imbalance in favor of Th2 cells provokes adipose tissue dysfunction and insulin resistance [33]. AGE-RAGE signaling pathway is also one of the predicted pathways for NRON target genes. This pathway contributes to the incidence and progression of type 2 diabetes by accumulating advanced glycation end products, which can activate the receptor for AGEs and trigger damaging cellular responses that are associated with impaired glucose tolerance, insulin resistance, inflammation, and microvascular complications [34]. Another primary function is Tumor Necrosis Factor (TNF) signaling pathway, which has a major role in type 2 diabetes pathogenesis. A cytokine released by macrophages and adipocytes, TNF is linked to the growth of insulin resistance and glucose intolerance. TNF signaling pathway paves the way for the development of type 2 diabetes by provoking inflammation, insulin resistance, oxidative stress, and pancreatic β-cell dysfunction [35]. Circadian rhythm was also identified as a central related mechanism. Gubin et al., reported that the disruptions in the normal circadian rhythm can be linked to the development of type 2 diabetes as it regulates the timing of biological processes, including insulin sensitivity and glucose metabolism [36]. Mitophagy is also one of the predicted functions for the targeted genes. Hippo signaling pathway is another function that involves NRON target genes. It has been shown that the Hippo signaling pathway regulates pancreatic beta-cell growth, insulin secretion, and glucose homeostasis. Hence, the dysregulation of this pathway can lead to reduced beta-cell proliferation, insulin secretion, impaired glucose tolerance, and therefore, incidence of type 2 diabetes. This pathway is also involved in regulation of insulin signaling, and its dysregulation can prompt inflammation, oxidative stress, and apoptosis, all of which are contributing factors of Type 2 diabetes [37]. Our data displayed the role of NRON target genes in the estrogen and prolactin signaling pathways. These pathways are crucial in regulating glucose metabolism and insulin sensitivity and influence the risk of type 2 diabetes. Estrogen signaling improves glucose uptake and lipid metabolism, while prolactin signaling stimulates insulin secretion and improves glucose uptake. However, the dysregulation of these pathways, due to diminished estrogen levels and consistently elevated levels of prolactin, can give rise to insulin resistance and development of type 2 diabetes [38,39]. In this regard, thyroid hormone signaling pathway has also been observed. TH levels have been shown to play a crucial role in the development and progression of insulin resistance. Research shows that the TH signaling pathway can directly affect insulin signaling pathways and glucose metabolism. Suboptimal thyroid hormone levels have been linked to insulin resistance and may contribute to the incidence of type 2 diabetes [40].
Informed by our results and findings of previous studies, it can be concluded that lncRNA NRON and its target biomarkers including NFATc3, are fundamentally involved in the incidence and progression of type 2 diabetes, as well as glucose regulation in patients with T2DM, and are therefore potential therapeutic targets for this disorder.
The present study had a number of limitations. Firstly, further research with a larger sample size is warranted. Secondly, longterm complications in T2DM, such as diabetic foot ulcer, retinopathy, nephropathy, and coronary artery disease, were not investigated. Finally, a comprehensive evaluation of other regulatory targets of lncRNA NRON and their differentially expressed levels in patients with T2DM is recommended.
Conclusion
In conclusion, marked differences were noticed in the expression levels of lncRNA NRON and NFATc3 between subjects in T2DM group and healthy controls. Moreover, their expression was correlated with metabolic features. We also found several pivotal genes associated with T2DM are targeted by lncRNA NRON, which are involved in a number of diabetes-related pathways, and NFATc3 was introduced as a central protein. These findings can shed further light on lncRNA NRON and NFATc3 regulations in T2DM and their therapeutic functions.
Acknowledgments
The authors would like to thank all subjects for their participation in the present study. We also appreciate the support of the University of Guilan.
References
- Galicia GU, Benito VA, Jebari S, et al. Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci 2020; 21(17):6275.
- Yang F, Chen Y, Xue Z, et al. High-throughput sequencing and exploration of the lncRNA-circRNA-miRNA-mRNA network in type 2 diabetes mellitus. Biomed Res Int 2020; 20:2020.
- Farhadnejad H, Teymoori F, Asghari G, et al. The higher adherence to a healthy lifestyle score is associated with a decreased risk of type 2 diabetes in Iranian adults. BMC Endocr Disord. 2022;22(1):42.
- Bener A, Zirie M, Rikabi A . Genetics, obesity, and environmental risk factors associated with type 2 diabetes. Croat Med J. 2005; 46:302-307.
- Brunetti A, Chiefari E, Foti D. Recent advances in the molecular genetics of type 2 diabetes mellitus. World J Diabetes.2014; 5:128.
- Sekar D, Nallaswamy D, Lakshmanan G. Decoding the functional role of long noncoding RNAs (lncRNAs) in hypertension progression. Hypertens Res. 2020; 43(7):724-5.
- Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci .2015; 16(2):3251-66.
- Khan S, Masood M, Gaur H, et al. Long non-coding RNA: An immune cells perspective. Life Sci. 2021; 271:119152.
- Yang F, Qin Y, Wang Y, et al. LncRNA KCNQ1OT1 Mediates Pyroptosis in Diabetic Cardiomyopathy. Cell Physiol Biochem. 2018; 50:1230-1244.
- Sathishkumar C, Prabu P, Mohan V, et al. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018; 12:41.
- Zhang W, Zheng J, Hu X, et al. Dysregulated expression of long noncoding RNAs serves as diagnostic biomarkers of type 2 diabetes mellitus. Endocrine. 2019; 65:494-503.
- Yang F, Chen Y, Xue Z, et al. High-Throughput Sequencing and Exploration of the lncRNA-circRNA-miRNA-mRNA Network in Type 2 Diabetes Mellitus. Biomed. Res Int 2020; 8162524.
- Du M, Wang C, Yang L, et al. The role of long noncoding RNA Nron in atherosclerosis development and plaque stability. iScience. 2020; 25:103978.
- Sharma S, Findlay GM, Bandukwala HS, et al. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci USA. 2011; 108:11381-11386.
- Xiong T, Huang C, Li J, et al. LncRNA NRON promotes the proliferation, metastasis and EMT process in bladder cancer. J Cancer. 2020; 11:1751-1760.
- Cai Y, Yao H, Sun Z, et al. Role of NFAT in the Progression of Diabetic Atherosclerosis. Front cardiovasc med. 2021; 8:635172.
- Heit JJ, Apelqvist AA, Gu X, et al. Calcineurin/NFAT signalling regulates pancreatic β-cell growth and function. Nature. 2006; 443:345-349.
- Tong Y, Zhang Z, Cheng Y, et al. Hypoxia-induced NFATc3 deSUMOylation enhances pancreatic carcinoma progression. Cell Death Dis. 2022; 13:1-13.
- Dai C, Hang Y, Shostak A, et al. Age-dependent human β cell proliferation induced by glucagon-like peptide 1 and calcineurin signaling. J Clin Investig. 2017; 127:3835-3844.
- Keller MP, Paul PK, Rabaglia ME, et al. The transcription factor Nfatc2 regulates β-cell proliferation and genes associated with type 2 diabetes in mouse and human islets. PLoS genetics. 2016; 12:e1006466.
- Corkum CP, Ings DP, Burgess C, et al. Immune cell subsets and their gene expression profiles from human PBMC isolated by Vacutainer Cell Preparation Tube (CPT™) and standard density gradient. BMC Immunol. 2015; 16:48.
- Zhang Y, Wu H, Wang F, et al. Long non-coding RNA MALAT1 expression in patients with gestational diabetes mellitus. Int J Gynaecol Obstet. 2018; 140:164-169.
- Carter G, Miladinovic B, Patel AA, et al. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 2015; 4:102-107.
- Huang X, Tan J, Li Y, et al. Expression of LncRNA KCNQ1Ot1 in diabetic nephropathy and its correlation with MEK/ERK signaling pathway. Am J Transl Res. 2022; 14:1796-1806.
- Ji X, Sun J, Wang Z. High level of lncRNA NR2F1-AS1 predict the onset and progression of diabetic retinopathy in type 2 diabetes. Exp Eye Res. 2022; 219:109069.
- Hu L, He F, Luo Y, et al. Reduced Compensatory β-Cell Proliferation in NFATc3-Deficient Mice Fed on High-Fat Diet. Exp Clin Endocrinol Diabetes. 2021; 129:651-660.
- Li J, Zhang Q, Jiao H. LncRNA NRON promotes M2 macrophage polarization and alleviates atrial fibrosis through suppressing exosomal miR-23a derived from atrial myocytes. J Formos Med Assoc. 2021; 120:1512-1519.
- Sun F, Guo Z, Zhang C, et al. LncRNA NRON alleviates atrial fibrosis through suppression of M1 macrophages activated by atrial myocytes. Biosci Rep 39(11):BSR20192215.
- Wang X, Chang X, Zhang P, et al. Aberrant expression of long non-coding RNAs in newly diagnosed type 2 diabetes indicates potential roles in chronic inflammation and insulin resistance. Cell Physiol Biochem. 2017; 43:2367-2378.
- Jiang H, Lou P, Chen X, et al. Deregulation of lncRNA HIST1H2AG-6 and AIM1-3 in peripheral blood mononuclear cells is associated with newly diagnosed type 2 diabetes. BMC Medical Genom. 2021; 14:149.
- Shiny A, Regin B, Mohan Vet al. Coordinated augmentation of NFAT and NOD signaling mediates proliferative VSMC phenotype switch under hyperinsulinemia. Atherosclerosis. 2016; 246:257-266.
- Chen J, Ning C, Mu J, et al. Role of Wnt signaling pathways in type 2 diabetes mellitus. Mol Cell Biochem. 2021; 476:2219-2232.
- Mahlangu T, Dludla PV, Nyambuya TM, et al. A systematic review on the functional role of Th1/Th2 cytokines in type 2 diabetes and related metabolic complications. Cytokine. 2020; 126:154892.
- Wu XQ, Zhang DD, Wang YN, et al. AGE/RAGE in diabetic kidney disease and ageing kidney. Free Radical Biol Med.2021; 171:260-271.
- Akash MSH, Rehman K, Liaqat A. Tumor necrosis factor‐alpha: Role in development of insulin resistance and pathogenesis of type 2 diabetes mellitus. J Cell Biochem. 2018; 119:105-110.
- Gubin D, Nelaeva A, Uzhakova A, et al. Disrupted circadian rhythms of body temperature, heart rate and fasting blood glucose in prediabetes and type 2 diabetes mellitus. Chronobiol Int. 2017; 34:1136-1148.
- Kilanowska A, Ziółkowska A. Apoptosis in type 2 diabetes: Can it be prevented? Hippo pathway prospects. Int J Mol Sci. 2022; 23:636.
- De Paoli M, Zakharia A, Werstuck GH. The role of estrogen in insulin resistance: A review of clinical and preclinical data. Am Jof Pathol. 2021; 191:1490-1498.
- Manshaei N, Shakibaei F, Fazilati M, et al. An investigation of the association between the level of prolactin in serum and type II diabetes. Diabetes Metab Syndr. 2019; 13:3035-3041.
- De Vito P, Candelotti E, Ahmed R, et al. Role of thyroid hormones in insulin resistance and diabetes. Imm Endoc Metab Agent. 2015;15:86-93.
 ) Healthy
control, (
) Healthy
control, ( ) T2DM patients.
) T2DM patients.