Research Article - Microbiology: Current Research (2023) Volume 7, Issue 3
Status of begomovirus on soybean crop and virus management strategies.
Khushbu Bathri, Sunil Kumar Snehi*Department of Microbiology, Barkatullah University, Bhopal, M.P, India
- *Corresponding Author:
- Sunil Kumar Snehi
Department of Microbiology
Barkatullah University, Bhopal, M.P, India
E-mail: sunilsnehi@gmail.com
Received: 25-May-2023, Manuscript No. AAMCR-23-103942; Editor assigned: 29-May-2023, PreQC No. AAMCR-23-103942(PQ); Reviewed: 12-Jun-2023, QC No. AAMCR-23-103942; Revised: 17-Jun-2023, Manuscript No. AAMCR-23-103942(R); Published: 24-Jun-2023, DOI:10.35841/aamcr-7.3.146
Citation: Bathri K, Snehi SK. Status of begomovirus on soybean crop and virus management strategies. J Micro Curr Res. 2023; 7(3):146
Abstract
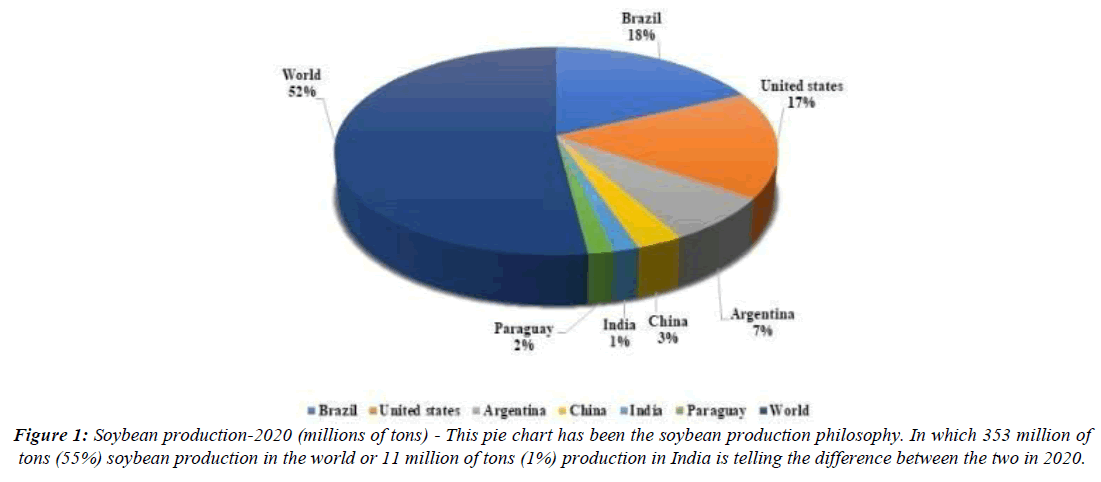
Soybean is a very important crop from the economic point of view of the farmers, but it has been seen that due to egomovirus (Family Geminiviridae) there is lots of damage to soybean cultivation. Soybean is widely cultivated throughout the country in which the total production of soybean in 2020 was 353million tons, with Brazil accounting for 66% of the production in the United State, Maharashtra, and Madhya Pradesh alone account for 89% of total soybean production in India, followed by Rajasthan, Andhra Pradesh, Karnataka, Chhattisgarh, and Gujarat with 11% production in these states. Several strategies have been adopted to protect soybean cultivars from begomovirus, including conventional and non-conventional management strategies for virus containment and are described in this article.
Keywords
Begomovirus, Soybean, Soybean mosaic disease, Management strategies.
Introduction
Soybean is a vital and nutritious crop [1]. Soybean is classified as a member of the order Fabales, the family Fabaceae, the subfamily Faboidae, and the genus Glycine. Glycine is divided into two subgenera: glycine, which contains 16 perennial species, and Soja Moench F.J. Herm, which contains two annual species, Glycine soja Siebold and Zucc (2n=40) and Glycine max (L.) Merrill (2n=40) [2]. Because of its numerous applications, this crop has been dubbed the "Golden Bean" or "Miracle Crop" of the twentieth century [3]. Apart from high-quality protein and oil, soybean contains a variety of therapeutic components such as lactose-free fatty acids, antioxidants such as vitamins C, K, and D, and folic acid, vitamins of the B complex group such as nicotinic acid (23 g/g), pantothenic acid, and folic acid, thiamine (12 g/g), pyridoxine (8 g/g), riboflavin (3.5 g/g), and biotin (0.7 g/g), as well as isoflavones such as genistein and daidzein [4]. In 2020, global soybean production was over 353 million tonnes, with Brazil and the United States accounting for 66% of the total (Figure 1). Production has increased dramatically worldwide since the 1960s, but particularly in South America since the 1980s, when a cultivar that grew well in low latitudes was developed [5]. The industry's rapid growth has been fueled primarily by large increases in global demand for meat products, particularly in developing countries such as China, which accounts for more than 60% of imports [6]. Madhya Pradesh and Maharashtra dominate soybean production in India, accounting for 89% of total output. The remaining 11% production is contributed by state-wise soybean cultivation statistics in lakh MT Rajasthan (10.558), Andhra Pradesh (2.840), Karnataka (2.470), Chhattisgarh (1.560), and Gujarat (0.930) [7].
The losses caused by soybean viruses are difficult to estimate due to interactions between soybean cultivars, time of infection, and virus strain. Nearly 67 viral soybean diseases have been identified worldwide, with at least 27 of them considered a threat to soybean cultivation [8]. Alfalfa Mosaic Virus (AMV; Alfamovirus), Bean Yellow Mosaic Virus (BYMV; Potyvirus), Cow Pea Mild Mottle Virus (CPMMV; Carlavirus), Groundnut Bud Necrosis Virus (GBNV; Tospovirus), Mungbean Yellow Mosaic India Virus (MYMIV; Begomovirus), Soybean Mosaic Virus (SMV; Potyvirus), Tobacco Ring Spot Virus (TRSV; Nepovirus), and Tobacco Streak Virus (TSV; Ilarvirus) [9]. New viral diseases such as Soybean dwarf virus, Tobacco streak virus, and Soybean vein necrosis virus can reduce yield even further. Soybean vein necrosis virus (Bunyavirales: Tospoviridae) was first identified in Tennessee in 2008 [10], Tobacco streak virus [11], Cucumber mosaic virus [12,13], Tomato spotted wilt virus [14,15]. Soybean chlorotic spot virus [16], Nepovirus [17], Soybean yellow shoot virus [18], African cassava mosaic virus [19-21], Cotton leaf curl Kokhran virus and Tomato leaf curl Karnataka virus [22,23], papaya leaf crumple virus [24]. One of the legume crops, soybean, is a good host for Begomovirus and a whitefly vector because of which there is widespread crop loss. There are numerous begomovirus species that infect legumes, including soybean being cited (Table 1).
| S.No. | Virus Group | Family | Genus | Species |
|---|---|---|---|---|
| 1. | Single-stranded DNA | Geminiviridae | Begomovirus | Abutilon mosaic virus African soybean dwarf virus Bean dwarf mosaic virus Bean golden mosaic virus Horse gram yellow mosaic virus Mung bean yellow mosaic virus Rhynchotia mosaic virus Sida mottle virus Soybean crinkle leaf virus Soybean chlorotic spot virus Tomato leaf curl New Delhi virus |
| 2. | Double-stranded DNA | Caulimovidae | Caulimovirus | Soybean chlorotic mottle virus |
Table 1. - Begomovirus infecting soybean
International status of begomovirus on soybean
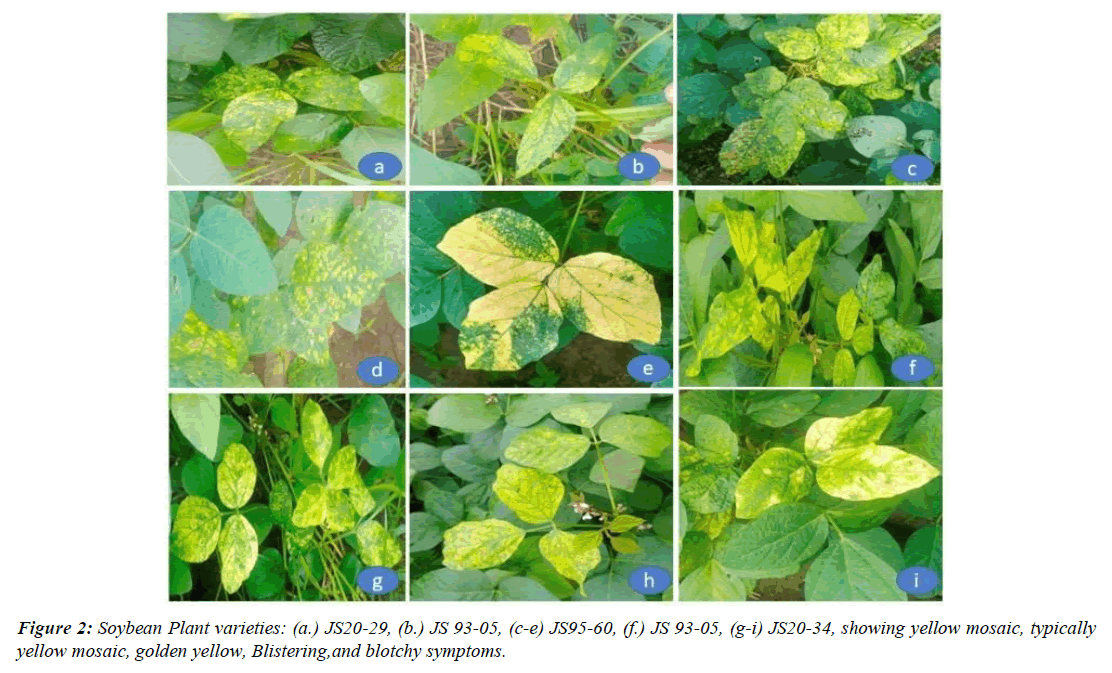
A Begomovirus linked to Soybean Leaf Curling and Chlorosis in Sinaloa, Mexico, is related to Pepper golden mosaic virus [25]. Tomato Leaf Curl New Delhi Virus, a bipartite begomovirus, infecting soybean for the first time in Faisalabad, Pakistan [26]. In Sinaloa, Mexico, a novel strain of Rhynchosia golden mosaic virus was identified from soybeans and weeds [27]. Soybean Crinkle Leaf Virus Complete Nucleotide Sequence and Genome Organization, Soybean Crinkle Leaf Virus (SCLV) causes soybean crop damage in various parts of Thailand [28]. The presence of African cassava mosaic virus in a mosaic disease of soybean in Nigeria is the first report [29]. A soybean chlorotic spot virus is a new begomovirus that infects soybean in Brazil [15]. In Nigeria, two novel 'legumoviruses', Soybean chlorotic blotch virus and Soybean mild mottle virus (genus Begomovirus), are naturally infected soybeans and causing symptoms such as trifoliate leaves and moderate mosaic [30]. In central Brazil, three separate begomoviruses relate to soybean, causing symptoms such as yellow and golden mosaic, chlorotic mottling, blistering, leaf deformation, and dwarfing [31]. Begomovirus genetic diversity and phylogeography in Pakistani legumes [32]. Alternate hosts of African cassava mosaic virus and East African cassava mosaic Cameroon virus in Nigeria [33]. All these viruses are shown in the Table 2 and virus symptoms Figure 2.
| S No. | Virus Name | Abbreviation | Accssetion Number | Symptoms | Country/Region | Reference |
|---|---|---|---|---|---|---|
| 1. | African cassava mosaic virus | ACMV | EU367500 | Yellow mosaic and mottling | Nigeria | [29] |
| 2. | African cassava mosaic virus | ACMV | EU685325 | Mosaic and mottling | Nigeria | [33] |
| 3. | Bean golden mosaic virus | BGMV | FJ665283 | Yellow and golden mosaic, chlorotic mottling, blistering, leaf distortion and dwarfing | Santo Antonio de Goia´s, State of Goia´s, Brazill | [31] |
| 4. | Okra mottle virus | OMoV | FJ686695 | Yellow and golden mosaic, chlorotic mottling, blistering, leaf distortion and dwarfing | Brazil | [31] |
| 5. | Pepper golden mosaic virus | PepGMV | AY905553 | Yellowing, leaf curling, crumpling and sunted growth | Sinaloa, Mexico | [25] |
| 6. | Rhynchosia golden mosaic virus | RhGMV | DQ347950 | Yellowing,Curled and stunting leaves | Mexico | [27] |
| 7. | Soybean crinkle leaf virus | SCLV | AB050781 | Twisting or curling of leaves and development of veinal enations on the under-surface of the leaves | Thailand | [28] |
| 8. | Soybean chlorotic spot virus | SoCSV | JX122965 | Leaf distorsion, blisteringInterveinal chlorosis, mosaic and golden mosaic | Brazil | [15] |
| 9. | Soybean chlorotic blotch virus | SbCBV | NC_014141 | Trifoliate leaves | Nigeria | [30] |
| 10. | Soybean chlorotic bloch virus | SbCBV | GQ472985 | Bright yellowing with blotchy | International Institute of Agriculture (IITA) farm in Ibadan, Oyo State, Nigeria | [30] |
| 11. | Soybean chlorotic bloch virus | SbCBV | GQ472986 | Foliar symptoms | Nigeria | [30] |
| 12. | Soybean mild mottle virus | SMMV | NC_014140 | Trifoliate leaves | Nigeria | [30] |
| 13. | Soybean mild mottle virus | GQ472984 | Mild mottle | Nigeria | [30] | |
| 14. | Sida micrantha mosaic virus | SiMMV | FJ686693 | Yellow and golden mosaic,chlorotic mottling,blistering, leaf and dwarfing | Brazil | [31] |
| 15. | Tomato leaf curl New Delhi virus | ToLCNDV | KX827599 | leaf curling, vein thickening and leaf yellowing | Faisalabad,Pakistan | [26] |
| 16. | Tomato leaf curl Pakistan betasatellite | ToLCPKV | AM922485 | Typical yellow mosaic | Pakistan | [32] |
Table 2. International status of begomovirus on soybean
National status of begomovirus on soybean
Yellow Mosaic Virus (YMV) infestation has exacerbated the issues for soybean farming in India. Natural infection of viruses such as Cotton leaf curl Kokhran virus, Tomato leaf curl Karnataka virus, and Papaya leaf crumple virus (Genus: Begomovirus) on soybean has been reported. Soybean plants with chlorosis, mosaic mottling, and necrosis of the leaves, petiole, stem, and the pods were gathered from the Maharashtra areas of Jalna, Beed, and Osmanabad. Mungbean Yellow Mosaic India Virus-resistant soybean cultivars promote viral RNA degradation earlier than susceptible cultivars [34]. Molecular analysis of two soybean-infecting begomoviruses from India, as well as evidence for recombination among South-East Asian legume-infecting begomoviruses on Mungbean yellow mosaic virus and Soybean mosaic virus [35]. Molecular evidence of a Meghalaya, India, mid-hills-based isolation of the mungbean yellow mosaic virus containing a recombinant DNA B component [36]. Ageratum enation virus full nucleotide sequence and an alphasatellite infecting a novel host Glycine max in Palampur India [37]. All these viruses are shown in the Table 2 and virus symptoms in Figure 2.
Management of Begomovirus
Plant virus diseases management
Plant virus diseases cause damage to plant growth, causing billions of dollars in damage to the crops of the world's farmers every year [38]. Plant viruses are summarized as sub microscopic units. Plant viruses are enveloped by nucleic acid proteins, which are said to be responsible for replicating inside a cell, and viruses cannot survive without a living cell, therefore it is considered as the link between the living. Plants are intracellular obligate parasites, which cannot survive without living cells/tissue. Plant viruses cannot be controlled by any chemical methods. Due to which farmers and gardeners are all worried. It is difficult to control plant virus by direct methods, that is why atypical methods are adopted [39]. The main factors driving the growth of the virus are:
• The monocrops density and genetic diversity of the plant, which makes it more susceptible to pathogens and organisms.
• Trade in the world of living plants and germplasm and which takes the virus host and vector to new areas.
• Viruses that rapidly grow in population, evolve, and adapt [40]. Conventional and nonconventional methods are used for the prevention of plant virus diseases [41].
Conventional Measures
Conventional measures are based on best practices, which can control virus diseases to a great extent. Virus diseases can be controlled to a great extent by using to know the virus, its diagnostic methods tissue culture is adopted, and for this virus removal materials such as routing, intercropping, avoidance is used.
Culture control and eliminat the weed host
For a long time, weeds are considered effective for spreading plant virus infection in the plant world and these weeds that grow throughout the year cause great damage to the crops, important in these weeds are Croton bonplandium, Acalypha Indica, Malvasrtumcoromandalianum, Eclipta alba, Ageratum conozoides, LaunaeaLaunaea procumbens, Jatropha gossypifolia, Cocciniagrandis, Nicotiana plumbaginifolia, Sorghum vulgare, Parthenium hysterophorus, Physalis minima, Sida cordifolia, and Sonchus oleraceus, these weeds have the highest risk of spreading bagomovirus and are mostly found in different places of India. According to, 13 species of weeds are found in India. And according to there are 18 weed species, due to which Bagomovirus spreads in crops. For the control of weeds growing throughout the year, weeds can be controlled to a great extent by strategies like a greenhouse, plantation planning, plant-to-plant distance and white mulch, and the productivity of the crops can be increased [43,44].
Quarantine regulation
Quarantine is a very useful method for the control of the pathogen. Plant and seed health testing is called quarantine. The European and Mediterranean Plant Protection Organization for Plant Grow Practices has been created, which has created a certification scheme for disease-free herbaceous plants, with the main goal of ensuring that plant growing practices meet their health standards. This certification scheme protects both nursery workers selling vegetatively propagated plant material and those buying nursery products, so the certification scheme was established. It mainly involves selection of health planting material, virus testing, microbial propagation, and testing for genetic fidelity.
Development and breeding for resistance
One approach is to develop different varieties of plants resistant to vector pests using techniques to control virus diseases [45]. An important example is the following is the resistance to tomato - infection that is represented by begomovirus from Solanum pimpinellifolium, Solanum peruvianum, Solanum chilense, and Solanum habrochaites [46]. A partially dominant major resistance gene, Ty-1, was produced by introgression from S. chilense and mapped to the short arm of chromosome 6 [47]. A major resistance Quantitative Trait Locus (QTL) derived from S. Pimpinellifolium (Hirsute-INRA) was mapped to a different position on chromosome 6 (TG153-CT83) [48]. The mapped a dominant resistance gene, Ty-2, in S. Habrochaites-derived line H24, to the short arm of chromosome 11. A partially dominant major gene, Ty-3, derived from S. chilense (LA2779 and LA1932), was mapped to chromosome 6 [49]. The Ty-3 introgression derived from LA2779 was found to be longer and linked to Ty-1. However, studies on fine mapping and characterization demonstrated that Ty-1 and Ty-3 are allelic and code for an RNA-dependent RNA polymerase [50]. An additional gene, Ty-4, was mapped to the long arm of chromosome 3. While Ty-3 has a major effect that accounts for 60% of the variation in symptom severity, Ty-4 accounts for only 16% of the variation [51]. A recessive resistance gene (Ty-5) has been identified on chromosome 4 in the lines derived from cultivar Ty king [52], which is suspected to be like the Ty-5 locus that accounts for more than 40% of the variation [53]. Most of these resistance sources are known to support virus replication. However, the level of virus accumulation is lower than the levels in susceptible cultivars. It is well established that the virus level in tomato lines carrying Ty-1/Ty-3 is ,10% of the level found in susceptible cultivars. Similarly, a low level of virus accumulation and a positive correlation between virus level and disease severity were found in Ty-2carrying lines [54].
Non-conventional Measures
Some methods of reducing plant viruses have been described previously. Begomovirus has some drawbacks of its own. The natural resistance to many viruses remains to be known. By raising resistant plants through genetic engineering, viral diseases can be saved or reduced to a great extent. The Steady introduction of genes of interest to plants from different organisms to control viruses is one of the most important developments in the lack of progress in agriculture. One is technology, which includes the use of agricultural chemicals to control pests and modern plant breeding, hybrid seed production and agricultural mechanization [55]. The Agrobacterium tumefaciens vector, the first transgenic engineering plant was produced from foreign genes, which was the first in the control of viruses. It was a great achievement. Genetic engineering produced the first virus resistance in the tobacco plant. The resulting transgenic gene from the plant's coat protein gene of the Tobacco mosaic virus was inserted into the plant. Both stalks about the presence of foreign lines [56]. The first breakthrough was engineering herbicide resistance and resistance using the coat with the Bacillus thuringiensis toxin gene [57].
Pathogen derived resistance
Together in 1986, Powell - Abel and his co-workers used a genetic engineering method to protect plants from viral diseases that catalyse genes in plants to generate resistant transgenic plants [58], in which 30 different groups of different viruses are used, for which different genes have been used and engineering resistance has been achieved [59-67]. This method is very important for genetic engineering. And proved to be very helpful in controlling virus diseases in crops inducing resistance of pathogens by mutation called Pathogen-Derived Resistance (PDR) with genes derived from the pathogen's genome which was first described by [68]. And a generalized concept expanded in 1985 by Sanford and Johnston is the binding of cross production with PDR by which a symptomless stain of the virus can cross plants [69]. Altered viral deriving genes are used to disrupt steps. Viral life cycles such as uncoating, replication, cell to cell or long distance or vector mediated transmission are where the genes of the virus are acquired or used [70]. There is coat putty, movement protein replicase (Rep) gene, antisense RNA, satellite RNA and defective interfering genes. CP is the most used transgene.
The coat protein works during the life span of the virus. It forms a shell, in which it strengthens its grip with the DNA of the virus, and protects it. In plant RNA represents and intermediate in a self-binding, nuclear targeting, or systematic movement [71]. It is extremely important for coat protein to be transmitted by insect vectors. In transgenic plants, the virus is caused by the expression of coat protein genes [72].
The protein intermediate is affected by the resistance coat protein gene in which a copy of a trans gene is inserted through which the trans gene passes, followed by a high level of transcription and translation of the protein, and is a medium level of resistance. It was considered like what happened earlier. The cross-protection coat protein hinders the uncoating of the virion and reduces or inhibits both cell-to-cell virus infection and spread, such as TMB, alpha mosaic virus to potato virus. All these viruses have transgenic coat proteins in transgenic plants, in terms of security. The coat protein messenger RNA and the coat protein were not resistant to infection [73-79].
The movement from cell to cell is the movement of plant virus to the outside plants, which is called movement protein mediate resistance. Externally, the movement protein in plants binds with the plasmodesmata and facilitates virus movement in the cell [80,81]. Previously used for engineering resistance to TMB in tobacco, a modified movement protein was prepared as a form of transgene resistance, which binds to the plasmodesmata side based on competition between the virus-encoded movement protein and the previously made inactive movement protein [82-87]
Conclusion
Viral pathogenicity on soybean is a serious economic threat that has a wide impact on growth and yield without being widely recognized. Even through, reports are accumulating about soybean infections with plant viruses, there is a lack of effective disease management of crops. The only way to succeed in plant disease management is having proper identification strategies to detect the viral pathogens early and accurately. Molecular assays for plant viruses are the detection method that has huge potential of accuracy. Hence, the development of new-molecular methods based on viral genomes facilitates the identification and diagnosis of plant viruses easily.
Conflict of Interest
The author declares that there is no conflict of interest.
References
- Campos RE, Bejerman N, Nome C, et al. Bean yellow mosaic virus in soybean from Argentina. J Phytopathol. 2014;162(5):322-5.
- Singh RJ, Hymowitz T. Soybean genetic resources and crop improvement. Genome. 1999;42(4):605-16.
- Latha TK, Malathi VG, Haokip BD, et al. First occurrence of bean common mosaic virus in soybean [Glycine max] from India. Australas Plant Dis Notes. 2017;12:1-3.
- Mathur S. Soybean the Wonder Legume. Beverage Food World. 2004; 31:61-62.
- De Maria M, Robinson EJ, et al. Global soybean trade-the geopolitics of a bean.
- Cattelan AJ, Dall’Agnol A. The rapid soybean growth in Brazil. OCL. 2018;25(1):D102.
- The soybean processors association of India. This evaluation report has been prepared by the Agriculture Division of FICCI
- Maroof MS, Tucker DM, Tolin SA. Genomics of viral–soybean interactions. InGene Genomics Soy. 2008;293-319.
- Sandra N, Tripathi A, Lal SK, et al. Molecular and biological characterization of soybean yellow mottle mosaic virus severe strain infecting soybean in India. 3 Biotech. 2021;11:1-2.
- Tzanetakis I, We R, Newman M, et al. Soybean vein necrosis virus: a new threat to soybean production in Southeastern United States? InPhytopathology 2009;99(6):131.
- Hong JS, Ohnishi S, Masuta C, et al. Infection of soybean by Cucumber mosaic virus as determined by viral movement protein. Arch Virol. 2007;152:321-8.
- Hosseinzadeh H, Nasrollanejad S, Khateri H. First report of cucumber mosaic virus subgroups i and ii on soybean, pea, and eggplant in iran. Acta virologica. 2012;56(2):145.
- Nischwitz C, Mullis SW, Gitaitis RD, et al. First report of tomato spotted wilt virus in soybean (Glycine max) in Georgia. Plant Dis. 2006;90(4):524.
- Yoon YN, Jo Y, Cho WK, et al. First report of Tomato spotted wilt virus infecting soybean in Korea. Plant Dis. 2018;102(2):461.
- Coco D, Calil IP, Brustolini OJ, et al. Soybean chlorotic spot virus, a novel begomovirus infecting soybean in Brazil. Arch Virol. 2013;158:457-62.
- Yasmin T, Nelson BD, Hobbs HA, et al. Molecular characterization of a new soybean-infecting member of the genus Nepovirus identified by high-throughput sequencing. Arch Virol. 2017;162:1089-92.
- Figueira AD, Geraldino-Duarte PS, Pinzon Nunez AM, et al. Characterization of soybean yellow shoot virus, a new member of the family Potyviridae infecting soybean plants in Brazil. Plant Dis. 2019;103(6):1172-80.
- Pietersen G, Garnett HM. A survey for the viruses of soybeans (Glycine max) in the transvaal, South Africa. Phytophylactica. 1990;22(1):35-40.
- Alabi OJ, Ogbe FO, Bandyopadhyay R, et al. Alternate hosts of African cassava mosaic virus and East African cassava mosaic Cameroon virus in Nigeria. Arch Virol. 2008;153:1743-7.
- Mgbechi-Ezeri JU, Alabi OJ, Naidu RA, et al. First report of the occurrence of African cassava mosaic virus in a mosaic disease of soybean in Nigeria. Plant Dis. 2008;92(12):1709.
- Raj SK, Khan MS, Snehi SK, et al. A yellow mosaic disease of soybean in northern India is caused by Cotton leaf curl Kokhran virus. Plant Dis. 2006;90(7):975.
- Raj SK, Khan MS, Snehi SK, et al. First report of Tomato leaf curl Karnataka virus infecting soybean in India. Plant Pathol. 2006;55(6).
- Jaidi M, Srivastava A, Kumar S, et al. First report of natural occurrence of Papaya leaf crumple virus on soyabean in India. New Dis Rep. 2015;32(1):15.
- Jamil N, Rehman A, Hamza M, et al. First report of Tomato leaf curl New Delhi virus, a bipartite begomovirus, infecting soybean (Glycine max). Plant Dis. 2017;101(5):845.
- Quintero-Zamora E, Barbosa-Jasso MP, Leyva-López NE, et al. A Begomovirus Associated with Leaf Curling and Chlorosis of Soybean in Sinaloa, Mexico is Related to Pepper golden mosaic virus. Plant Dis. 2006;90:109.
- Jamil N, Mansoor S, Amin I. First Report of Tomato Leaf Curl New Delhi Virus, a bipartite begomovirus, infecting Soybean (Glycine max). Plant disease. 2016;101:845-845.
- Mauricio-Castillo JA, Mendez-Lozano J, Perea-Araujo L, Arguello-Astorga G.R. A new strain of Rhynchosia golden mosaic virus isolated from soybean and weeds in Sinaloa, Mexico. Plant Disease. 2006; 90:972
- Samretwanich K, Kittipakorn K, Chiemsombat P, et al. Complete nucleotide sequence and genome organization of soybean crinkle leaf virus. J Phytopathol. 2001;149(6):333-6.
- Mgbechi-Ezeri JU, Alabi OJ, Naidu RA, et al. First report of the occurrence of African cassava mosaic virus in a mosaic disease of soybean in Nigeria. Plant Dis. 2008;92(12):1709.
- Alabi OJ, Lava Kumar P, Mgbechi-Ezeri JU, et al. Two new ‘legumoviruses’(genus Begomovirus) naturally infecting soybean in Nigeria. Arch Virol. 2010;155:643-56.
- Fernandes FR, Cruz AR, Faria JC, et al. Three distinct begomoviruses associated with soybean in central Brazil. Arch Virol. 2009;154:1567-70.
- Ilyas M, Qazi J, Mansoor S, et al. Genetic diversity and phylogeography of begomoviruses infecting legumes in Pakistan. J Gener Virol. 2010;91(8):2091-101.
- Yadav RK, Shukla RK, Chattopadhyay D. Soybean cultivar resistant to Mungbean Yellow Mosaic India Virus infection induces viral RNA degradation earlier than the susceptible cultivar. Virus Res. 2009;144(1-2):89-95.
- Girish KR, Usha R. Molecular characterization of two soybean-infecting begomoviruses from India and evidence for recombination among legume-infecting begomoviruses from South-East Asia. Virus Res. 2005;108(1-2):167-76.
- Mumford R, Macarthur R, and Boonham N. The role and challenges of new diagnostic technology in plant biosecurity. Food Secur. 2016;8:103–109.
- Kumar S, Kumari A, Raj R, et al. Management of viral diseases of crops. Appl Plant Virol. 2020;575-92.
- Rubio L, Galipienso L, Ferriol I. Detection of plant viruses and disease management: Relevance of genetic diversity and evolution. Front Plant Sci. 2020;11:1092.
- Banerjee A, Umbrey Y, Yadav RM, et al. Molecular evidence of an isolate of mungbean yellow mosaic India virus with a recombinant DNA B component occurring on mungbean from mid-hills of Meghalaya, India. Virus Dis. 2018;29:68-74.
- Snehi SK, Raj SK, Prasad V and Singh V. Recent Research Findings Related to Management Strategies of Begomoviruses. Plant Pathol Microb. 2015; 6:273
- Khan MS, Raj SK, Singh BP. Some weeds as new hosts of geminivirus as evidenced by molecular probes. Indian J Plant Pathol. 2003;21:82-5.
- Singh J, Sohi AS, Mann HS, et al. Studies on whitefly, Bemisia tabaci (Genn.) transmitted cotton leaf curl disease in Punjab. J Insect Sci. 1994;7(2):194-8.
- Valand GB, Muniyappa V. Epidemiology of tobacco leaf curl virus in India. Ann Appl Bio. 1992;120(2):257-67.
- Waterworth P, RP K. Thermotherapy and aseptic bud culture of sugarcane to facilitate the exchange of germplasm and passage trough quarantine. Plant Dis Rep. 1978;62:72-776
- Golino DA, Savino V, Martelli GP, et al. Certification and international regulation of planting material. Compend Grape Dis. APS Press, St. Paul. 2005.
- Valkonen J. Virus disease control in plants using natural and engineered resistance, and some considerations regarding biosafety. Current. 1998;51-5.
- Ji Y, Schuster DJ, Scott JW. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol Breed. 2007;20:271-84.
- Zamir D, Ekstein-Michelson I, Zakay Y, et al. Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor Appl Genet. 1994;88:141-6.
- Chagué V, Mercier JC, Guenard M, et al. Identification of RAPD markers linked to a locus involved in quantitative resistance to TYLCV in tomato by bulked segregant analysis. Theor Appl Genet. 1997;95:671-7.
- Hanson PM, Bernacchi D, Green S, et al. Mapping a wild tomato introgression associated with tomato yellow leaf curl virus resistance in a cultivated tomato line. J Am Soc Hort Sci. 2000;125(1):15-20.
- Verlaan MG, Hutton SF, Ibrahem RM, et al. The Tomato Yellow Leaf Curl Virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD class RNA-dependent RNA polymerases. PLoS Genet. 2013;9(3):e1003399
- Ji Y, Scott JW, Schuster DJ, et al. Molecular mapping of Ty-4, a new tomato yellow leaf curl virus resistance locus on chromosome 3 of tomato. J Am Soc Hort Sci. 2009;134(2):281-8.
- Hutton SF, Scott JW, Schuster DJ. Recessive resistance to Tomato yellow leaf curl virus from the tomato cultivar Tyking is located in the same region as Ty-5 on chromosome 4. Hort Sci. 2012;47(3):324-
- Anbinder I, Reuveni M, Azari R, Paran I, Nahon S et al. Molecular dissection of Tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor. Appl. Genet. 2009; 119:519-530
- Barbieri M, Acciarri N, Sabatini E, et al. Introgression of resistance to two Mediterranean virus species causing tomato yellow leaf curl into a valuable traditional tomato variety. J Plant Pathol. 2010:485-93.
- Gasser CS, Fraley RT. Genetically engineering plants for crop improvement. Sci. 1989;244(4910):1293-9.
- Abel PP, Nelson RS, De B, et al. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Sci. 1986;232(4751):738-43.
- Horsch RB, Fry JE, Hoffmann NL, et al. A simple and general method for transferring genes into plants. Science. 1985;227(4691):1229-31.
- Gonsalves D, Slightom JL. Coat protein-mediated protection: analysis of transgenic plants for resistance in a variety of crops. InSeminars in Virol. 1993; 4(6):397-405.
- Lomonossoff GP. Pathogen-derived resistance to plant viruses. Annu Rev Phytopathol. 1995;33(1):323-43.
- Pappu HR, Niblett CL, Lee RF. Application of recombinant DNA technology to plant protection: molecular approaches to engineering virus resistance in crop plants. World J Microbio Biotechnol. 1995;11:426-37.
- Varma A. Application of biotechnology in plant pest management: current status and future prospects. Proceedings of regional expert consultation on application of biotechnology in plant pest management. FAO, RAP Public. 1997:21-66.
- Prins M, Goldbach R. The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbio. 1998;6(1):31-5.
- Reimann-Philipp U. Mechanisms of resistance: Expression of coat protein. Plant Virol Prot: From Virus Isol Transg Resist. 1998:521-32.
- Bendahmane M, Beachy RN. Control of tobamovirus infections via pathogen-derived resistance. Adv Vir Res. 1999;53:369-86.
- Jain RK, Varma A. Biotechnological management of viral diseases of plants. Plant diseases. Pointer Publish. 2000:1-20.
- Callaway A, Giesman-Cookmeyer D, Gillock ET, et al. The multifunctional capsid proteins of plant RNA viruses. Ann Rev Phytopathol. 2001;39(1):419-60.
- Varma A, Jain RK, Bhat AI. Virus resistant transgenic plants for environmentally safe management of viral diseases.
- Hamilton RI. In: Viruses. Plant disease: An advanced treatise. Academic Press, NY 1980; 5:279.
- Sanford JC, Johnston SA. The concept of parasite-derived resistance-deriving resistance genes from the parasite's own genome. J Theor Bio. 1985;113(2):395-405.
- Grumet R. Development of virus resistant plants via genetic engineering. Plant Breed Rev. 2010;12:47-79.
- Ivanov KI, Mäkinen K. Coat proteins, host factors and plant viral replication. Cur Opi Virol. 2012;2(6):712-8.
- Ng JC, Liu S, Perry KL. Cucumber mosaic virus mutants with altered physical properties and defective in aphid vector transmission. Virol. 2000;276(2):395-403.
- Beachy RN, Loesch-Fries S, Tumer NE. Coat protein-mediated resistance against virus infection. Ann Rev Phytopathol. 1990;28(1):451-72.
- Wisniewski LA, Powell PA, Nelson RS, et al. Local and systemic spread of tobacco mosaic virus in transgenic tobacco. Plant Cell. 1990;2(6):559-67.
- Abel PP, Nelson RS, De B, et al. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science. 1986;232(4751):738-43.
- Loesch‐Fries LS, Merlo D, Zinnen T, et al. Expression of alfalfa mosaic virus RNA 4 in transgenic plants confers virus resistance. EMBO Journal. 1987;6(7):1845-51.
- Hemenway C, Fang RX, Kaniewski WK, et al. Analysis of the mechanism of protection in transgenic plants expressing the potato virus X coat protein or its antisense RNA. EMBO J. 1988;7(5):1273-80.
- Hayakawa T, Zhu Y, Itoh K,et al.Genetically engineered rice resistant to rice stripe virus, an insect-transmitted virus. Proc Natl Acad Sci.1992;89(20):9865-9.
- Powell PA, Stark DM, Sanders PR, et al. Protection against tobacco mosaic virus in transgenic plants that express tobacco mosaic virus antisense RNA. Proc Natl Acad Sci. 1989;86(18):6949-52.
- Wolf S, Lucas WJ, Deom CM, et al. Movement protein of tobacco mosaic virus modifies plasmodesmatal size exclusion limit. Sci. 1989;246(4928):377-9.
- Deom CM, Schubert KR, Wolf S, et al. Molecular characterization and biological function of the movement protein of tobacco mosaic virus in transgenic plants. Proc Natl Acad Sci. 1990;87(9):3284-8.
- Malyshenko SI, Kondakova OA, Nazarova JV, et al. Reduction of tobacco mosaic virus accumulation in transgenic plants producing non-functional viral transport proteins. J Gen Virol. 1993;74(6):1149-56.
- Lapidot M, Gafny R, Ding B, et al. A dysfunctional movement protein of tobacco mosaic virus that partially modifies the plasmodesmata and limits virus spread in transgenic plants. Plant J. 1993;4(6):959-70.
- Alabi OJ, Ogbe FO, Bandyopadhyay R, et al. Alternate hosts of African cassava mosaic virus and East African cassava mosaic Cameroon virus in Nigeria. Arch Virol. 2008; 153:1743-7.
- Kumar NA, Narasu ML, Zehr UB, et al. Molecular characterization of Tobacco streak virus causing soybean necrosis in India.
- Sharma S, Singh L, Roshan P, et al. Complete nucleotide sequence of Ageratum enation virus and an alphasatellite infecting a new host Glycine max in India. Jo Phytopathol. 2016;164(7-8):554-7.
- Soybean production in 2019, Crops/World regions/Production quantity (from pick lists). United Nations, Food and Agriculture Organization, Statistics Division, FAOSTAT. 2019. Retrieved February 8, 2021.
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref
Indexed at, Google Scholar, Cross ref