- Biomedical Research (2013) Volume 24, Issue 4
Short communication: Effects of a 17-beta estradiol analogue on gene expression and morphology in a breast epithelial adenocarcinoma cell line: A potential antiproliferative agent
Michelle Helen Visagie1, Barend André Stander1, Lyn-Marie Birkholtz2, Annie Margaretha Joubert1*1Department of Physiology, University of Pretoria, Private Bag X323, Arcadia, 0007, South Africa
2Department of Biochemistry, University of Pretoria, Private Bag X20 Hatfield, Pretoria, 0028, South Africa
- *Corresponding Author:
- Annie Joubert
Department of Physiology
University of Pretoria, Bag X 323
Arcadia, 0007, South Africa
Accepted August 19 2013
Citation: Visagie MH, Stander BA, Birkholtz L-M, Joubert AM. Short communication: Effects of a 17-beta estradiol analogue on gene expression and morphology in a breast epithelial adenocarcinoma cell line: A potential antiproliferative agent. Biomedical Research 2013; 24 (4): 525-530.
Abstract
2-Methoxyestradiol is a 17-beta estradiol derivative with antitumor activity. Due to low bioavailability, a new estrogen analogue was in silico-designed. The latter, a sulphamoylated analogue of 2-methoxyestradiol, was evaluated for its potential antiproliferative effect by means of expression microarray analysis and transmission electron microscopy in the highly metastatic breast adenocarcinoma (MDA-MB-231) cell line. Data indicate changes in gene expression pertaining to induction of apoptosis and autophagy as types of cell death, arrest of the cell cycle, and impairment of the cytoskeleton and induction of reactive oxygen species. Transmission electron microscopy confirmed morphological characteristics of apoptosis and autophagy. Altered cell cycle- and cytoskeletal-related gene expression profiles demonstrated that the estradiol analogue acts as an antimitotic agent in this highly metastatic breast cell line. Data also showed that the newly designed estrogen analogue exerts an antiproliferative effect in this cancer cell line culminating in both apoptosis and autophagy as type of cell death paving the way for further investigations into its potential as anticancer agent.
Keywords
Apoptosis, microarray, autophagy, antimitotic, cell cycle
Introduction
Microtubules are viewed as an ideal target for chemotherapeutic agents due to its crucial role during the formation of the dynamic spindle apparatus in mitosis [1]. 2-Methoxyestradiol is a naturally occurring derivative of β-estradiol that exerts microtubule disruption and antitumor activity. However, 2-methoxyestradiol failed to advance to United State Food and Drug Administration approval due to limited biological accessibility [3]. Reports indicated that estrogen sulphamate analogues featuring a 2-methoxy, 2-ethyl, or 2-methyl sulphanyl group possess optimal antiproliferative activity [4-5]. This potent antitumor activity is due to their ability to interact with sulphatase, carbonic anhydrase and proteins such as tubulin [5].
The addition of a 3-O-sulphamate group allows for resistance to inactivating conjugation of the C-3 hydroxyl group, which estradiol derivatives are usually subjected to [4]. In addition, the estrogen 3-O-sulphamates are highly reversible inhibitors of carbonic anhydrase II. The latter is most likely responsible for the high bioavailability of the sulphamoylated analogues since reversible uptake by red blood cells and interaction with carbonic anhydrase II ensures transiting the liver without undergoing first pass metabolism [4-6]. This study focused on the potential antiproliferative effect of a novel 2-methoxyestradiol analogue namely (8R, 13S, 14S, 17S)-2-Ethyl-13-methyl- 7, 8, 9, 11, 12,13, 14, 15, 16, 17-decahydro-6Hcyclopenta[ a]phenanthrane-3, 17-diyl bis(sulphamate) (EMBS) on gene expression and morphology on the triple negative adenocarcinoma (MDA-MB-231) cell line. (Figure 1).
Materials and Methods
The MDA-MB-231 is a tumorigenic estrogen receptor negative breast epithelial carcinoma cell line that is derived from a metastatic adenocarcinoma site and is commercially available from Microsep (Pty) ltd, Johannesburg (South Africa). All required reagents of cell culture analytical grade were purchased from Sigma (St. Louis, United States of America). The RNeasy Mini Kit was provided by Southern Cross Biotechnology Pty (Ltd) (Cape Town, South Africa). Agilent’s Human 1A Oligo Microarray slides, Agilent Gene Expression Hybridization Kit, Agilent Expression Wash Buffer Kit and the Agilent RNA spike-in kit was provided by Agilent Technologies Pty (Ltd) (Palo Alto, States of America). The Quick AMP labelling kit was bought from Chemetrix (Midrand, South Africa). The Nanodrop ND- 1000 spectrophotometer (Nanodrop Technologies, Wilmington, USA) utilized for spectrophotometrical quantification of ribonucleic acid (RNA) and deoxyribonucleic acid (DNA) and the Axon Genepix 400B scanner (Molecular Devices, Sunnyvale, USA) utilized for microarray slide scanning was supplied by the ACGT Microarray Facility of the University of Pretoria (www.microarray.up.ac.za; Gauteng, South Africa).
EMBS was synthesized by iThemba Pharmaceuticals (Pty) Ltd. (Modderfontein, South Africa) since this compound was not commercially available [6]. A stock solution of EMBS dissolved in dimethyl sulphoxide (DMSO) was prepared with a concentration of 10 mM and was stored at 4°C. The vehicle control sample composed of DMSO (v/v) and growth medium. Cells were exposed to 0.4 μM EMBS for 24 h as prior experiments conducted in our laboratory (Department of Physiology, University of Pretoria, South Africa) indicated successful antiproliferative activity at this concentration (data not shown). Cells were grown and maintained in 25 cm2 tissue culture flasks in a humidified atmosphere at 37°C and 5% CO2 and cultured in Dulbecco’s Minimum Essential medium eagle (DMEM), supplemented with 10% heat-inactivated fetal calf serum (56°C, 30 min), 100 U/mL penicillin G, 100 μg/mL streptomycin and fungizone (250 μg/L).
Whole human genome gene expression microarrays
Agilent’s Human 1A Oligo 60-mer Microarray (V2) 44 k slides with more than 41 000 60-mer oligonucleotide human genes and transcripts were employed to study global expression changes on the MDA-MB-231 cell line induced after 24 h treatment of EMBS according to the method described by Stander, et al (2013) [7]. A dyeswop methodology with three biological replicates was employed in order to remove the effects of dye-bias on statistical analyses and make the genomic information statistically relevant. Slides were scanned with the Axon Genepix 4000B Scanner (Molecular Devices, USA) provided by the ACGT Microarray Facility at the University of Pretoria.
Differentially expressed upregulated- and downregulated genes were established by means of the log fold change of a particular gene (LogFC) value. LogFC values provide the difference by means of the log2 transformed gene expression data (data not shown). Positive logFC values demonstrate the logarithmic foldness of upregulation and negative logFC values indicate the logarithmic foldness of downregulation in gene expression.
Transcripts with a B-value of greater than zero were considered statistically significantly differentially expressed. The B-statistic is defined as log-odds that that transcript is differentially expressed and a B-statistic of zero corresponds to a 50/50 chance that the transcripts are differentially expressed. Statistically significant genes involved in the major cell death pathways (apoptosis and autophagy) and antiproliferative relevant categories related to possible anticancer activity were grouped in specific categories.
Transmission electron microscopy
Transmission electron microscopy (TEM) was conducted to demonstrate effects on EMBS on cell morphology to visualize characteristics of apoptosis and autophagy. TEM was performed according to Visagie and Joubert (2010) [3]. Samples were viewed with a JOEL JEM 2100F transmission electron microscope (Electron Microscopy Unit, University of Pretoria, South Africa).
Statistics
All experiments were conducted in triplicate. Qualitative data was supplied by means of TEM and gene expression microarrays provided quantitative data. Scanning and data analysis of microarrays were conducted according to the method described by Stander, et al (2013) [7].
Results
Whole human genome gene expression microarrays
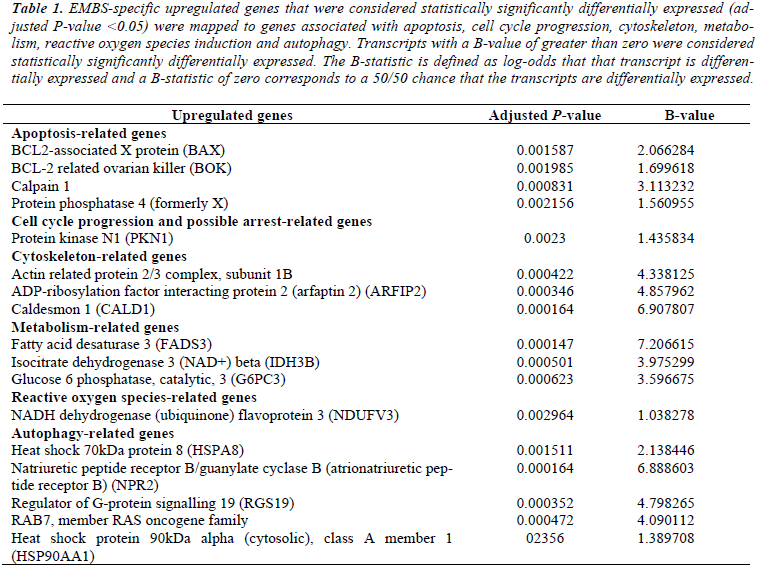
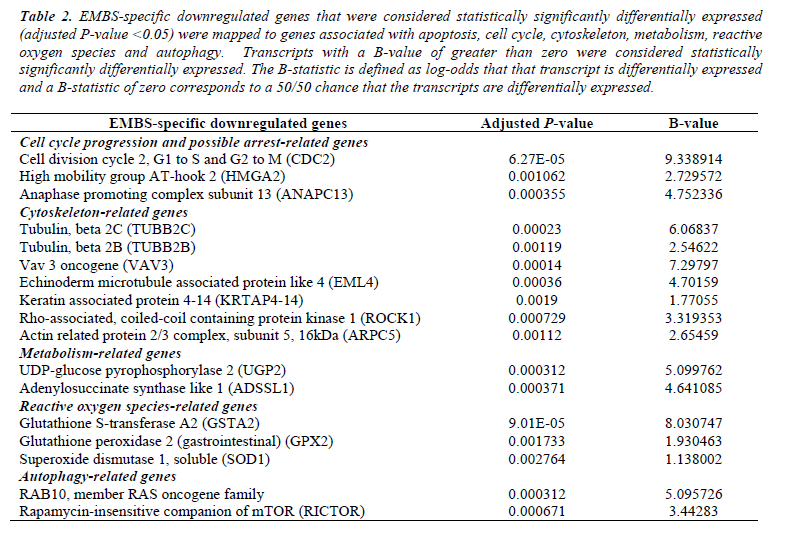
Microarray studies (whole human genome) were performed to study the effects of EMBS on gene expression in the highly metastatic MDA-MB-231 cell line. EMBS caused changes in gene expression levels demonstrating cell death induction via apoptosis and autophagy. Genes that were considered statistically significantly differentially expressed (adjusted P-value <0.05) in treated cells were mapped to genes associated with cell growth, apoptosis, autophagy, cell cycle and DNA repair and oxidative stress. Transmission electron microscopy confirmed morphological characteristics of apoptosis and autophagy (Table 1 and Table 2).
Table 1: EMBS-specific upregulated genes that were considered statistically significantly differentially expressed (adjusted P-value <0.05) were mapped to genes associated with apoptosis, cell cycle progression, cytoskeleton, metabolism, reactive oxygen species induction and autophagy. Transcripts with a B-value of greater than zero were considered statistically significantly differentially expressed. The B-statistic is defined as log-odds that that transcript is differentially expressed and a B-statistic of zero corresponds to a 50/50 chance that the transcripts are differentially expressed.
Table 2: EMBS-specific downregulated genes that were considered statistically significantly differentially expressed (adjusted P-value <0.05) were mapped to genes associated with apoptosis, cell cycle, cytoskeleton, metabolism, reactive oxygen species and autophagy. Transcripts with a B-value of greater than zero were considered statistically significantly differentially expressed. The B-statistic is defined as log-odds that that transcript is differentially expressed and a B-statistic of zero corresponds to a 50/50 chance that the transcripts are differentially expressed.
Transmission electron microscopy
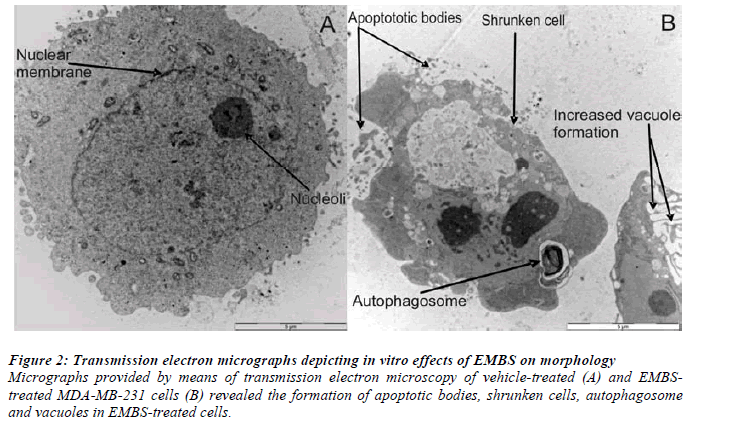
Transmission electron microscopy (TEM) demonstrated morphological characteristics of cell death and confirmed antiproliferative effects of EMBS on these breast cancer cells. TEM illustrated the presence of apoptotic bodies, shrunken cells, autophagosome and vacuoles when compared to vehicle-treated control cells (Figure 2).
Figure 2: Transmission electron micrographs depicting in vitro effects of EMBS on morphology.
Micrographs provided by means of transmission electron microscopy of vehicle-treated (A) and EMBStreated
MDA-MB-231 cells (B) revealed the formation of apoptotic bodies, shrunken cells, autophagosome
and vacuoles in EMBS-treated cells.
Discussion
This study is the first to investigate the antiproliferative effect of a novel sulphamoylated 2-methoxyestradiol derivative of 17-beta estadiol on gene expression and morphology in an estrogen receptor-negative, highly metastatic breast adenocarcinoma cell line. Gene expression microarray data suggested that EMBS causes its antiproliferative effect by possible induction of apoptosis and autophagy. Altered gene expression analysis indicated that EMBS also causes impairment of cytoskeletal structure, -metabolism, induction of reactive oxygen species and arrest in cell cycle progression. Characteristics of both apoptosis and autophagy as types of cell death were observed by transmission electron microscopy.
Altered cell cycle-specific gene expression signifies that EMBS exposure results in G2/M arrest. The possible antimitotic activity of EMBS is supported by the downregulated TUBB2C, TUBB2C, ELM4 and CALD1, which demonstrate that EMBS targets the actin and tubulin for conformational changes.
The induction of autophagy is indicated by the altered expression of the autophagy-related genes. Rab7 is involved in endocytosis and enhancement of the fusion between autophagosome and lysosomes during autophagy and thus increased Rab7 expression indicates the increased presence of autophagy [8]. Rab10 plays a role in vesicle transport and reports indicate that RAB10 inhibition results in increased autophagy and this may be the mechanism by which dietary restriction stimulates autophagy [9]. The mTORC2 protein rapamycininsensitive companion of mTOR (rictor) regulates autophagy and inhibition of the mTORC results in autophagy induction in the non-small H383 cell lung cancer cell line [10].
In conclusion, this gene expression study indicates that EMBS induces apoptosis and autophagy, exerts antimitotic activity and increases reactive oxygen species generation paving the way for future investigations into its potential as antiproliferative, and ultimately, an anticancer agent.
Declaration of competing interests
The authors declare there are not any competing interests.
Contribution by authors
MHV and AMJ conducted the project design. MHV conducted the experiments and data analysis and compiled the manuscript. ABS assisted in the global DNA microarray experiments and the microarray statistical analysis. LMB and AMJ supervised this project, revised the manuscript proofread the manuscript and acted as editors. AMJ managed the funding acquisition from various grants. All authors have contributed and approved the final manuscript.
Acknowledgements
This study was supported by grants from the Cancer Association of South Africa, the Struwig Germeshuysen Trust, RESCOM (Research Council of the University of Pretoria), National Research Foundation and The Medical Research Council.
References
- Stanton RA, Gernert KM, Nettles JH, Aneja R. Drugs that target dynamic microtubules: a new molecular per- spective. Med Res Rev 2011; 31: 443-481.
- Risinger AL, Westbrook CD, Encinas M, Mülbaier M, Schultes CM, Wawro S, Lewis JD, Janssen B, Giles FJ, Mooberry SL. ELR5010444, a novel microtubule dis- ruptor with multiple mechanisms of action. Pharmacol Exp Ther 2011; 6: 652-660.
- Visagie MH, Joubert AM. In vitro effects of 2- methoxyestradiol-bis-sulphamate on cell numbers, membrane integrity, morphology and possible induc- tion of apoptosis and autophagy in a non-tumorigenic breast epithelial cell line. Cell Mol Biol Lett 2010; 15: 564-581.
- Leese LP, Jourdan FL, Gaokroger K, Mahon MF, Newman SP, Foster PA, Stengel C, Regis-Lydi S, Di Fiore A, De Simone G, Supuran CT, Purohit A, Reed MJ, Potter BVL. Structure-activity relationship of C-17 cyano-substituted estratriene as anticancer agents. J Med Chem 2008; 51: 1295-1308.
- Jourdan F, Leese MP, Dohle W, Hamel E, Ferrandism E, Newman SP, Purohit A, Rees MJ, Potter BVL. Synthesis, antitubulin, and antiproliferative SAR of analogues of 2-methoxyestradiol-3- 17-O, O-bis- sulfamate. J Med Chem 2010; 53: 2942-2951.
- Stander A, Joubert F, Joubert A. Docking, synthesis, and in vitro evaluation of antimitotic estrone analogues. Chem Biol Drug Des 2011; 77: 173-181.
- Stander BA, Joubert F, Yu C, Sippel KH, McKenna R, Joubert AM. Signaling pathways of ESE-16, an antimitotic and anhydrase estradiol analog, in breast cencer cells. PlosOne 2013; 8: e53853.
- Lin WJ, Yang CY, Li LL, Hsiang Y, Chen KW, Lin YC, Lin CC, Lin CH. Lysosomal targeting of phafin mediated by Rab7 induces autophagosome formation. Biochem Biophys Res Commun 2012; 417: 35-42.
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A Role for Autophagy in the Extension of Lifespan by Dietary Restriction in C. elegans. PLoS Genet 2008; 4: e24.
- Sini P, James D, Chresta C, Guichard S. Simultaneous inhibition of mTORC1 and mTORC2 by mTOR kinase inhibitor AZD8055 induces autophagy and cell death in cancer cells. Autophagy 2010; 6: 553-554.