Research Article - Archives of General Internal Medicine (2023) Volume 7, Issue 5
Resistance and susceptible prevelance study of Pseudomonas aeruginosa
Sonalika Mohapatra1,2*1Department of Microbiology, Ashwini Hospital, Cuttack India
2Department of Microbiology, Centurion University, Bhubaneswar, India
- *Corresponding Author:
- Sonalika Mohapatra
Department of Microbiology
Ashwini Hospital, Cuttack India
E-mail: sonasalony782@gmail.com
Received: 25-Sept-2023, Manuscript No. AAAGIM-23-114406; Editor assigned: 28-Sept-2023, PreQC No. AAAGIM-23-114406(PQ); Reviewed: 12-Oct-2023, QC No. AAAGIM-23-114406; Revised: 17-Oct-2023, Manuscript No. AAAGIM-23-114406(R); Published: 24-Oct-2023, DOI:10.35841/aaagim-7.5.193
Citation: Mohapatra S. Resistance and susceptible prevelance study of Pseudomonas aeruginosa. Arch Gen Intern Med. 2023;7(5):193
Abstract
Pseudomonas aeruginosa (P. aeruginosa) is a gram negative bacterium that continues to be a major cause of opportunistic nosocomial infections, causing around 9-10% of hospital infections. It is hard to treat because of intrinsic resistance of the species and its resistance to multiple groups of antibiotics including β-lactams, aminoglycosides and fluoroquinolones. This study was undertaken to determine the prevalence of P. aeruginosa and its susceptibility pattern isolated from pus, urine, blood etc. samples at Ashwini Hospital, Cuttack. Resistant Pseudomonas aeruginosa isolates are one of the major causes of both hospital-acquired infections (HAIs) and community-acquired infections (CAIs). However, management of P. aeruginosa infections is difficult as the bacterium is inherently resistant to many antibiotics. All P. aeruginosa isolates were tested for susceptibility to 11 commonly used antibiotics, and the newly introduced Double Locus Sequence Typing (DLST) scheme was implemented to elucidate the predominant clones. The tested P. aeruginosa isolates presented various resistant phenotypes, with Verona Integron-Mediated Metallo-β-lactamase (VIM-2) mechanisms being the majority, and a new phenotype, FEPR-CAZS, being reported for the first time in Greek isolates. DLST revealed two predominant types, 32-39 and 8-37, and provided evidence for intra-hospital transmission of the 32-39 clone in one of the hospitals. The results indicate that DLST can be a valuable tool when local outbreaks demand immediate tracking investigation with limited time and financial resources.
Keywords
Acute Cholecystitis, Most common finding, Ultrasound.
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a gram negative bacterium that continues to be a major cause of opportunistic nosocomial infections, causing around 9-10% of hospital infections. It is hard to treat because of intrinsic resistance of the species and its resistance to multiple groups of antibiotics including β-lactams, aminoglycosides and fluoroquinolones. This study was undertaken to determine the prevalence of P. aeruginosa and its susceptibility pattern isolated from pus samples.
Pseudomonas aeruginosa (P.aeruginosa) is one of the most common gram-negative microorganisms identified in the clinical specimens of hospital admitted patients. It is a commensal of human microflora in healthy people and is frequently isolated as an opportunistic pathogen in recurrent infections of hospitalized patients (Figure 1).
1. It can infect almost any external site or organ, and therefore,can be isolated from various body fluids such as sputum, urine, wounds, eye or ear swabs and from blood.
2. This organism is often hard to treat because of both the intrinsic resistance and acquired resistance i.e. mutations in chromosomal genes, to multiple groups of antimicrobial agents, including β-lactams, aminoglycosides and fluoroquinolones.
3. An increased resistance of P.aeruginosa to β-lactam drugs is because of production of metallo-beta-lactamases i.e. enzymes that efficiently hydrolyze all β-lactams.
4. The implication of these emerging resistance is in the successful treatment of infections caused by this bacteria cannot be overemphasized.
5. It causes infections in hospitalized patients particularly in burns, orthopedic related infections, respiratory diseases, catheterized and even immunosuppressed patients. Inherent resistance to many antimicrobial agents, contributes substantially to wound related morbidity and mortality worldwide.
Keeping in view the occurrence of Pseudomonas spp. in different habitat, its pathology and resistance to antibiotics, this study was aimed to isolate P.aeruginosa from pus samples and to determine its antibiotic susceptibility profile [1].
Isolates of Pseudomonas aeruginosa or Acinetobacter species that are resistant to all, or almost all, commercially available antibiotics are now prevalent worldwide. Typically, these strains are recovered from patients in intensive care units who have ventilator-associated pneumonia. "Panresistant" strains can be defined as strains that are resistant to all beta -lactam and quinolone antibiotics recommended as empirical therapy for ventilator-associated pneumonia. These strains are well adapted to the hospital environment--molecular epidemiological studies have frequently revealed that only 1 or 2 clones caused outbreaks in intensive care units. However, panresistant strains may also be selected by antibiotic use. Given the lack of antibiotic options to treat infection with panresistant strains, enhanced surveillance for these organisms is necessary at unit-specific, institutional, and national levels.
Review of Literature
Pseudomonas aeruginosa is a common pathogen that is implicated in a wide variety of nosocomial infections. Hospital mortality rates associated with P. aeruginosa bacteremia is reported to be >20% in recent studies. Inappropriate use of empirical antibiotic therapy has been identified as an independent contributor to the high hospital mortality rate of P. aeruginosa bacteremia.
Pseudomonas aeruginosa is a Gram-negative and ubiquitous environmental bacterium. It is an opportunistic human pathogen capable of causing a wide array of life-threatening acute and chronic infections, particularly in patients with compromised immune defense. It has been of particular importance since it is the main cause of morbidity and mortality in cystic fibrosis (CF) patients and one of the leading nosocomial pathogens affecting hospitalized patients while being intrinsically resistant to a wide range of antibiotics [2].
P. aeruginosa strains possess large genomes (~5–7 Mbp). Their metabolic capacity is extensive as exemplified by their ability to produce multiple secondary metabolites and polymers as well as their ability to use various carbon sources and electron acceptors [3]. The repertoire of P. aeruginosa genes which are substantially conserved suggest the highest proportion of regulatory genes and networks observed in known bacterial genomes and is foundational to respond and adapt to diverse environments. The ubiquitous presence of P. aeruginosa as well as its prevalence and persistence in clinical settings including intrinsic resistance to therapeutics are attributed to its extraordinary capability of survival by recruiting an arsenal of responsive mechanisms.
In the present review, we have attempted to summarize the diversity of these mechanisms causing the versatility of P. aeruginosa to adapt and thrive in unfavorable conditions particularly during pathogenesis. To this end, we will describe the clinical importance of P. aeruginosa followed by the well-characterized and most recent findings about key strategic adaptation mechanisms including quorum sensing (QS), motility-sessility switch, biofilm formation, antibiotic resistance mechanisms, adaptive radiation for persistence, stringent response and persisters, and the CRISPR-Cas systems [4]. Recent findings on adaptive mechanisms will be set into context of the overall physiology of P. aeruginosa by also depicting on future research needs.
Clinical Importance
The CF patients suffer from a multisystem disease due to inheritable genetic defects in the CF trans membrane conductance regulator (CFTR) gene. However, the recurrence of bacterial infections in the abnormal mucus layers is the main cause of morbidity and mortality of CF patients. The CFTR regulator is responsible for regulating the transport of electrolytes and chloride across epithelial cell membranes to maintain normal mucus properties and homeostasis. Therefore, the loss of function of the CFTR protein results in abnormally thick, dehydrated and sticky mucus layers in the lung. Hence, the CF patients are largely susceptible to respiratory infections by P. aeruginosa from infancy. When they are under a year old, almost 30% of CF infants can acquire initial P. aeruginosa strains from the environment leading to acute infections. This rate increases to about 50% by the age of 3 years while mucoid phenotypes causing chronic infections have been reported emerging at the age of 3 to16 years (median of 13 years) [5]. P. aeruginosa will adapt to CF airways and persist as overwhelming, predominant and ineradicable infections to the end of patients' life in almost 70% of adults.
Furthermore, P. aeruginosa is also largely associated with hospital acquired infections including ventilator-associated pneumonia, central line-associated bloodstream infection, urinary catheter-related infection, and surgical/transplantation infections. The International Nosocomial Infection Control Consortium reported that P. aeruginosa nosocomial infections have become a worldwide healthcare issue. A cohort study reported that P. aeruginosa had the highest burden of healthcare-acquired infections in European intensive care units [6]. In the United States healthcare-associated P. aeruginosa infections were estimated to contribute to 51,000 cases each year. P. aeruginosa is prevalent in healthcare settings because it is a common companion of patients under medical care and also it can survive on abiotic and biotic surfaces such as medical equipment resisting disinfection methods while being transmissible from patient-to-patient.
P. aeruginosa infections are becoming more difficult to treat because this bacterium is naturally resistant to many antibiotics and the number of multidrug- and pan-drug-resistant strains is increasing worldwide. Strains have been reported which are resistant to almost all class of commonly used antibiotics including aminoglycosides, cephalosporins, fluoroquinolones, and carbapenems. In the United States about 13% of P. aeruginosa infections are caused by multidrug resistant strains [7].
P. aeruginosa utilizes sophisticated genotypic events to support various phenotypes and molecular mechanisms required for survival during pathogenesis and antibiotic treatment.
How can you avoid getting an infection?
Patients and caregivers should:
• keep their hands clean to avoid getting sick and spreading germs that can cause infections
• wash their hands with soap and water or use alcoholbased hand sanitizer, particularly before and after caring for wounds or touching a medical device
• remind healthcare providers and caregivers to clean their hands before touching the patient or handling medical devices
• allow healthcare staff to clean their room daily when in a healthcare setting
Healthcare providers should pay careful attention to recommended infection control practices, including hand hygiene and environmental cleaning (e.g., cleaning of patient rooms and shared equipment) to reduce the risk of spreading these germs to patients.
Healthcare facilities should have water management plans (see Reduce Risk from Water) that help ensure water quality and reduce the risk of exposure to potentially harmful germs like Pseudomonas aeruginosa [8].
Central Regulatory Role of Quorum Sensing (QS) for Virulence and Adaptation
Communication between individual cells using specific chemical signals is a well-known capability of bacteria and is called quorum sensing. Indeed, QS controls social behavior of bacteria by multiple interconnected signaling pathways. It allows bacterial communities to regulate a variety of biological processes important for bacterial adaptation and survival. Basically, this phenomenon relies on regulating the expression of specific sets of genes in response to a critical threshold of signaling molecules known as Autoinducers (AIs). QS will mediate population density dependent collective responses and is therefore beneficial for community survival. A study showed that cells' responses to QS signals and the corresponding gene expression profile is heterogeneous within a given community leading to increasing fitness and chance of survival.
During pathogenesis P. aeruginosa QS plays a critical role for survival and colonization by coordinating phenotypic alterations at early stages of infection i.e., after attachment. The progress of acute to chronic infection is critically influenced by QS-dependent gene expression. More than 10% of P. aeruginosa genes are regulated by QS. These genes are mainly involved in virulence factor production, motility, motility-sessility switch and biofilm development, antibiotic resistance mechanisms and the adjustment of metabolic pathways for stress responses. The role of QS in each physiological adaptation will be discussed below.
Molecular Mechanisms Underlying QS
Four main pathways of QS dependent signaling exist in P. aeruginosa. These constitute a hierarchal network mediating integration of multiple signals via cross-talk between the QS signaling pathways. The most recently discovered IQS signaling pathway is less understood and its integration and impact on gene expression still needs to be unraveled. It was previously proposed that the IQS molecule (an aeruginaldehyde) is the product of enzymatic activity of proteins encoded by ambBCDE genes, while new findings showed the IQS molecule is a byproduct of the pyochelin biosynthesis pathway and AmbBCDE proteins are responsible for the biosynthesis of the toxin L-2-amino-4-methoxy-trans- 3-butenoic acid (AMB).
Signs and symptoms
Pseudomonas infections frequently involve the following parts of the body, with corresponding symptoms and signs: Endocarditis: Fever, murmur, and positive blood culture findings; peripheral stigmata such as Roth spots, Jane way lesions, Osler nodes, splinter hemorrhages, and splenomegaly.
Bacteremia: Fever, tachypnea, and tachycardia; hypotension and shock; jaundice. Pseudomonas bacteremia produces distinctive skin lesions known as etyma gangrenous. Though bacteremia can be caused by a multitude of mechanisms, some more frequent causes are urinary tract infections and users of intravenous narcotics. Skin and soft tissue infections: Hemorrhagic and necrotic lesions, with surrounding erythema; subcutaneous nodules, deep abscesses, cellulitis, and fasciitis; in burns, black or violaceous discoloration or eschar.
Ecthyma gangrenosum lesions are hemorrhagic and necrotic, with surrounding erythema. These characteristic lesions are almost always caused by Pseudomonas infection and usually are found in the axilla, groin, or perianal area but may involve any part of body. Pseudomonas also has emerged as an important source of burn wound sepsis. Invasive burn wound sepsis is defined as the bacterial proliferation of 100,000 organisms per gram of tissue, with subjacent involvement of subjacent unburned tissue.
Pseudomonal burn wound infections appear black or as a violaceous discoloration or eschar. Systemic manifestations of burn wound sepsis may include fever or hypothermia, disorientation, hypotension, oliguria, ileus, and leukopenia [9].
Diagnosis
Depending on the nature of infection, an appropriate specimen is collected and sent to a bacteriology laboratory for identification. As with most bacteriological specimens, a Gram stain is performed, which may show Gram-negative rods and/or Depending on the nature of infection, an appropriate specimen is collected and sent to a bacteriology laboratory for identification. As with most bacteriological specimens, a Gram stain is performed, which may show Gram-negative rods and/or white blood cells? P. aeruginosa produces colonies with a characteristic "grape-like" or "fresh-tortilla" odor on bacteriological media. In mixed cultures, it can be isolated as clear colonies on MacConkey agar (as it does not ferment lactose) which will test positive for oxidase. Confirmatory tests include production of the blue-green pigment pyocyanin on cetrimide agar and growth at 42 °C. A TSI slant is often used to distinguish nonfermenting Pseudomonas species from enteric pathogens in faecal specimens (Figure 2).
When P. aeruginosa is isolated from a normally sterile site (blood, bone, deep collections), it is generally considered dangerous, and almost always requires treatment. However, P. aeruginosa is frequently isolated from nonsterile sites (mouth swabs, sputum, etc.), and, under these circumstances, it may represent colonization and not infection. The isolation of P. aeruginosa from nonsterile specimens should, therefore, be interpreted cautiously, and the advice of a microbiologist or infectious diseases physician/pharmacist should be sought prior to starting treatment. Often, no treatment is needed cells. P. aeruginosa produces colonies with a characteristic "grape-like" or "fresh-tortilla" odor on bacteriological media. In mixed cultures, it can be isolated as clear colonies on MacConkey agar (as it does not ferment lactose) which will test positive for oxidase. Confirmatory tests include production of the blue-green pigment pyocyanin on cetrimide agar and growth at 42 °C. A TSI slant is often used to distinguish nonfermenting Pseudomonas species from enteric pathogens in faecal specimens [10].
When P. aeruginosa is isolated from a normally sterile site (blood, bone, deep collections), it is generally considered dangerous, and almost always requires treatment.However, P. aeruginosa is frequently isolated from nonsterile sites (mouth swabs, sputum, etc.), and, under these circumstances, it may represent colonization and not infection. The isolation of P. aeruginosa from nonsterile specimens should, therefore, be interpreted cautiously, and the advice of a microbiologist or infectious diseases physician/pharmacist should be sought prior to starting treatment. Often, no treatment is needed.
Treatment
Many P. aeruginosa isolates are resistant to a large range of antibiotics and may demonstrate additional resistance after unsuccessful treatment. It should usually be possible to guide treatment according to laboratory sensitivities, rather than choosing an antibiotic empirically. If antibiotics are started empirically, then every effort should be made to obtain cultures (before administering the first dose of antibiotic), and the choice of antibiotic used should be reviewed when the culture results are available.
Due to widespread resistance to many common first-line antibiotics, carbapenems, polymyxins, and more recently tigecycline were considered to be the drugs of choice; however, resistance to these drugs has also been reported. Despite this, they are still being used in areas where resistance has not yet been reported. Use of β-lactamase inhibitors such as sulbactam has been advised in combination with antibiotics to enhance antimicrobial action even in the presence of a certain level of resistance. Combination therapy after rigorous antimicrobial susceptibility testing has been found to be the best course of action in the treatment of multidrug-resistant P. aeruginosa. Some next-generation antibiotics that are reported as being active against P. aeruginosa include doripenem, ceftobiprole, and ceftaroline. However, these require more clinical trials for standardization. Therefore, research for the discovery of new antibiotics and drugs against P. aeruginosa is very much needed.
Pseudomonas infections are treated with antibiotics. Unfortunately, many pseudomonas infections are becoming more difficult to treat. These bacteria have developed the ability to adapt and overcome antibiotics in their environment. This is called antibiotic resistance.
The increase in antibiotic resistance has made treating infections much more challenging. Pseudomonas infectionscan often develop resistance to multiple types of antibiotics. It can even sometimes develop resistance during the course of treatment.
It is important that your doctor selects an effective antibiotic. A doctor may send a specimen from a patient to a laboratory first for testing in order to be more certain. The laboratory will test the specimen to determine which antibiotic will work best.
Treatment may involve one or more of the following types of antibiotics: ceftazidime, ciprofloxacin (Cipro) or levofloxacin, gentamicin, cefepime, aztreonam, carbapenems, ticarcillin, ureidopenicillins (Figure 3)
Antibiotic resistance
One of the most worrisome characteristics of P. aeruginosa is its low antibiotic susceptibility, which is attributable to a concerted action of multidrug efflux pumps with chromosomally encoded antibiotic resistance genes (e.g., mexAB, mexXY, etc.) and the low permeability of the bacterial cellular envelopes.In addition to this intrinsic resistance, P. aeruginosa easily develops acquired resistance either by mutation in chromosomally encoded genes or by the horizontal gene transfer of antibiotic resistance determinants. Development of multidrug resistance by P. aeruginosa isolates requires several different genetic events, including acquisition of different mutations and/or horizontal transfer of antibiotic resistance genes. Hypermutation favours the selection of mutation-driven antibiotic resistance in P. aeruginosa strains producing chronic infections, whereas the clustering of several different antibiotic resistance genes in integrons favors the concerted acquisition of antibiotic resistance determinants. Some recent studies have shown phenotypic resistance associated to biofilm formation or to the emergence of small-colony variants may be important in the response of P. aeruginosa populations to antibiotics treatment [11].
Mechanisms underlying antibiotic resistance have been found to include production of antibiotic-degrading or antibiotic-inactivating enzymes, outer membrane proteins to evict the antibiotics and mutations to change antibiotic targets. Presence of antibiotic-degrading enzymes such as extended-spectrum β-lactamases like PER-1, PER-2, VEB-1, AmpC cephalosporinases, carbapenemases like serine oxacillinases, metallo-b-lactamases, OXA-type carbapenemases, aminoglycoside-modifying enzymes, among others have been reported. P. aeruginosa can also modify the targets of antibiotic action, for example methylation of 16S rRNA to prevent aminoglycoside binding and modification of DNA, or topoisomerase to protect it from the action of quinolones. P. aeruginosa has also been reported to possess multidrug efflux pumps systems that confer resistance against a number of antibiotic classes and the MexAB-OprM (Resistance-nodulation-division (RND) family) is considered as the most important. An important factor found to be associated with antibiotic resistance is the decrease in the virulence capabilities of the resistant strain. Such findings have been reported in the case of rifampicin-resistant and colistin-resistant strains, in which decrease in infective ability, quorum sensing and motility have been documented [12].
Prevention
Probiotic prophylaxis may prevent colonization and delay onset of Pseudomonas infection in an ICU setting. Immunoprophylaxis against Pseudomonas is being investigated. The risk of contracting P. aeruginosa can be reduced by avoiding pools, hot tubs, and other bodies of standing water; regularly disinfecting and/or replacing equipment that regularly encounters moisture (such as contact lens equipment and solutions); and washing one's hands often (which is protective against many other pathogens as well). However, even the best hygiene practices cannot totally protect an individual against P. aeruginosa, given how common P. aeruginosa is in the environment.
Thoroughly washing hands and cleaning equipment in hospitals can help prevent infection. Outside a hospital, avoiding hot tubs and swimming pools that are poorly cared for can help prevent infections. You should remove swimming garments and shower with soap after getting out of the water. Drying your ears after swimming can also help prevent swimmer’s ear.
There are several things you can do to prevent infection if you are recovering from a procedure or receiving a treatment in a hospital:
• Tell your nurse if any of your dressings become loose or look wet.
• Tell your nurse if you think any tubes of IV lines have come loose.
• Make sure you fully understand the treatment or procedure your doctor has requested for you.
If you have diabetes, make sure you discuss controlling your blood sugar with your doctor before your procedure.
Risk Factor
You can get pseudomonas in many different ways. It can grow on fruits and vegetables, so you could get sick from eating contaminated food. It also thrives in moist areas like pools, hot tubs, bathrooms, kitchens, and sinks.
The most severe infections occur in hospitals. Pseudomonas can easily grow in humidifiers and types of medical equipment -- catheters, for instance -- that aren’t properly cleaned. If health care workers don’t wash their hands well, they can also transfer the bacteria from an infected patient to you.
Your risk of pseudomonas infection also goes up if you:
• Have a wound from surgery
• Are being treated for burns
• Use a breathing machine, catheter, or other medical device
• Have diabetes or cystic fibrosis
• Have a disorder that weakens your immune system, such as HIV
Take medications that suppress your immune system, like those that treat cancer.
Statistical analysis
Statistical analysis was done by descriptive statistics using simple ratio and percentages (Table 1-3).
| Department | No. of isolates (N=57) | % age |
|---|---|---|
| Surgery | 19 | 33.3 |
| ICU | 13 | 22.8 |
| ENT | 8 | 14.1 |
| Medicine | 7 | 12.3 |
| Orthopedics | 4 | 7 |
| OBG | 4 | 7 |
| Emergency | 2 | 3.5 |
Table 1. Department wise distribution of P.aeruginosa isolates.
| Age group(in years) | No. of isolates (N=57) | % age |
|---|---|---|
| =20 | 7 | 12.3 |
| 21-30 | 11 | 19.3 |
| 31-40 | 5 | 8.8 |
| 41-50 | 12 | 21 |
| 51-60 | 4 | 7 |
| 61-70 | 12 | 21 |
| >70 | 6 | 10.6 |
Table 2. Age wise distribution of Pseudomonas aeruginosa isolates.
| Department | No. of MDR isolates(N=14) | % age |
|---|---|---|
| ICU | 6 | 42.8 |
| Surgery | 5 | 35.7 |
| OBG | 2 | 14.2 |
| Emergency | 1 | 7.3 |
Table 3. Department-wise distribution of MDR P.aeruginosa isolates.
Antibiotic Resistance Mechanisms
Indeed, the emergence of antibiotic resistant bacteria is a global health issue. Among identified notoriously multi-drug resistant (MDR) bacteria, P. aeruginosa has been introduced as a major concern with a growing threat to global health resulting in dramatically increasing prevalence of nosocomial and chronic infections. This is due to the extraordinary capacity of these bacteria to develop resistance against a wide range of antimicrobials through various molecular mechanisms which are often simultaneously present in clinical isolates. Although each resistance mechanism is related to a specific class of antibiotics, multiple mechanisms mediate variably resistance to each class of antibiotics. Furthermore, the contribution of each mechanism varies from country to country. Loss or reduced copy numbers of OprD and overproduction of active efflux pumps, AmpC β-lactamase and extended-spectrum β-lactamases have been mainly reported as main contributors to multi-drug resistance phenotypes of P. aeruginosa isolates [13].
Recent reviews have described the prevalence and contribution of each resistance mechanism to each class of antibiotics in detail. Here, we reviewed the most frequent and well-understood findings which are classified into intrinsic, acquired and adaptive mechanisms, and we provide an update on our understanding of how P. aeruginosa can survive antibiotic treatments.
Intrinsic Resistance Mechanisms
Like many Gram-negative bacteria, P. aeruginosa can be intrinsically resistant to particular antibiotics. Such intrinsic resistance mechanisms stem from the existence of genes in bacterial genome encoding inherent properties of cell structures and composition providing protection against toxic molecules and antimicrobials. It can also be contributed by the lack of susceptible sites which naturally exist in antibiotic sensitive species
However, hydrophilic antibiotics can enter cells by diffusing through membrane channels or porin proteins in a non-specific manner. As one of the intrinsic mechanisms, P. aeruginosa limits antibiotic entry by reducing the number of non-specific porin proteins and replacing them with specific or moreselective channels for taking up required nutrients resulting in lowered permeability to toxic chemicals. P. aeruginosa resistance to currently used broad-spectrum drugs such as carbapenems and cephalosporins is commonly caused by this adaptation. Many of the clinical strains of P. aeruginosa displaying resistance to carbapenems such as imipenem are deficient in the OprD porin which specifically facilitates the diffusion of basic amino acids, small peptides as well as carbapenems into the cell.
Active multidrug efflux pumps greatly contribute to antibiotic resistance observed in P. aeruginosa. The involved genes are ubiquitous in Gram-negative bacteria and they are located on the genome or plasmids. The multidrug efflux pumps are multi-protein complexes spanning the envelope of Gramnegative bacteria. They are responsible for expelling various toxic materials and a wide range of antimicrobials. Because of their broad substrate specificities they display resistance against different classes of antibiotics which are chemically unrelated [14].
P. aeruginosa possesses four well known active multidrug efflux pumps including MexAB-OprM, MexXY/ OprM(OprA), MexCD-OprJ, and MexEF-OprN. The gene sets encoding these systems are under different regulatory factors; therefore, the expression levels of these systems significantly differ under various conditions. The MexABOprM and MexXY/OprM(OprA) are the most clinically important sets due to their large prevalence in clinical strains and significant contribution to a wide range of antibiotic. The mexAB-oprM genes show a stable and constitutive expression in the cell guaranteeing a protective basal level production of the MexAB-OprM system to consistently expel a wide range of toxic molecules and antibiotics. Hence it mainly contributes to natural resistance to antibiotics. The mexXY-(oprA) genes show lower basal expression levels and are mainly induced in response to protein synthesis inhibitors that target the ribosomal machinery. Both mexCD-oprJ and mexEF-oprN genes are not typically expressed in wild-type strains or their expression is very low and they have been proposed not to contribute significantly to natural antibiotic resistance.
There are other forms of multidrug efflux pumps such as MexJK, MexGHI-OpmD, MexVW, MexPQ-OpmE, MexMN, and TriABC. They are not expressed in wild-type strains but may contribute to adaptive resistance against antibiotic or biocide agents when expressed in resistant strains.
On the other hand, they might play role in other physiological pathways as well. For example, The MexEF-OprN and MexGHI-OpmD sets can modulate QS systems by exporting the quinolone signaling molecule PQS reducing its cellular concentration resulting in the reduction of virulence factor production, which is presumably in favor of establishment of chronic infections. However, many of these mechanisms remain still unclear with regard to their connection with other physiological pathways and their clinical relevance [15].
Another player of intrinsic resistance and basal lower level antibiotic susceptibility in P. aeruginosa is the gene encoding an inducible β-lactamase (AmpC). Particularly, chromosomal expression and production of AmpC confers low level resistance to aminopenicillins and most cephalosporins because these antibiotics strongly induce the production of AmpC which consequently hydrolyzes these substrates. However, through adaptive or acquired resistance mechanisms AmpC can be overproduced, consequently conferring resistance to a wider range of antibiotics such as aminoglycosides and fluoroquinolones. These mechanisms will be further discussed later.
Acquired Resistance Mechanisms
P. aeruginosa can acquire resistance to antibiotics through mutation of intrinsic genes or horizontal acquisition from other bacteria through transferring plasmids carrying genetic materials encoding for antibiotic resistance. Contrary to intrinsic mechanisms, acquired resistance is related to antibiotic selection and this selective advantage occurs in the presence of antibiotic compounds leading to irreversible resistant population. Therefore, similar to intrinsic resistance, acquired resistance is stable too and it can be transmitted to progeny.
However, due to over-expression of resistance genes and transmissibility by plasmids, acquired resistance is a potent mechanism which confers resistance to a wide spectrum of antibiotics as well as leads to increased prevalence among clinical and environmental strains.
Boosted Antibiotic Resistance via Mutations
Intrinsic resistance genes are negatively or positively regulated by one or more regulatory mechanisms which confer a basal lower susceptibility of P. aeruginosa to a narrow spectrum of antibiotics. However, mutation in regulatory pathway could increase promoter activities resulting in unleashing gene expression and overproduction of protein products such as AmpC and multi-drug efflux pumps systems. Consequently, it causes higher level of resistance to antibiotics.
As a common mutational feature of P. aeruginosa isolates, resistant clinical mutants display a constitutive high level of AmpC production even in the absence of antibiotic inducers. This is mainly due to mutational inactivation of ampD (repressor of ampC) and specific point mutations of ampR, both encoding two regulatory proteins important in induction of the ampC gene. Consequently, it turns into a major cause of greater resistance to a wide range of antibiotics such as most of the β-lactams (e.g., ticarcillin and piperacillin) as well as monobactams, third-generation and fourth-generation cephalosporins. One study showed that 73% of tested clinical strains showed AmpC overproduction.
Several regulatory loci such as mexR, nalD, nalB, and nalC negatively control the expression of the mexAB-oprM operon in P. aeruginosa. On the other hand, various loss-of-function mutations in these loci derepress the expression of the mexABoprM operon leading to the overproduction of MexAB-OprM complex conferring a greater resistance to carbapenem antibiotics. Likewise, overproduction of other multidrug efflux pumps such as MexXY and MexCD-OprJ can occur via mutations in regulatory loci leading to unleashing gene expression and a greater resistance to a variety of antimicrobial agents [16].
Another clinically important and prevalent mutational alteration is attributed to OprD porin channel. This porin channel is localized in the outer membrane of P. aeruginosa and it is characterized as a carbapenem-specific porin. Therefore, loss or reduction of OprD can reduce permeability of the outer membrane to carbapenems. The emergence of resistance to imipenem and reduced susceptibility to meropenem has been reported upon the occurrence of oprD mutations. Genetic alteration in oprD can occur via nucleotide insertion or deletion and point mutations resulting in frameshift of the gene sequence, amino acid substitution, shortened putative loop L7 and premature stop codons. Furthermore, downregulation of oprD expression can be mediated by other regulatory factors such as MexT which itself concurrently upregulates mexEFoprN expression.
Additionally, fluoroquinolone resistance among P. aeruginosa isolates can be mediated by either mutational changes within the fluoroquinolone targets i.e., DNA gyrase (gyrA and gyrB) and/or topoisomerase IV (parC and parE) or overproduction of active or inducible efflux pumps.
Plasmid-Mediated Resistance
Bacterial plasmids serve a central role as a potent vehicle for acquiring resistance genes and subsequent delivery to recipient host. This is so-called horizontal gene transfer whereby genetic elements can be transferred between bacterial cells particularly via conjugation. Some resistance plasmids are broad host range which can be transferred among various species via bacterial conjugation, while narrow host range plasmids are transferred among a small number of cells from similar bacterial species. For example, plasmid RP1 can transfer resistance genes to most Gram-negative bacteria.
Plasmid-encoded antibiotic resistance confers resistance to different classes of antibiotics that are currently applied in frontline of clinical treatments such as β-lactams, fluoroquinolones and aminoglycosides. So far, P. aeruginosa resistance via horizontal gene transfer has been reported for the genes encoding β-lactam-hydrolyzing enzymes known as the extended-spectrum β-lactamases and the carbapenemases, aminoglycoside-modifying enzymes, 16S rRNA methylases resulting in high-level pan-aminoglycoside resistance.
The genes encoding extended-spectrum β-lactamases and carbapenemase are clinically important not only due to their hydrolyzing activity on a wide range of β-lactams such as carbapenems and extended-spectrum cephalosporins, but also for their worldwide prevalence. The global epidemiology of carbapenem-resistant P. aeruginosa was recently analyzed by Hong et al. They reported that the geographical prevalence of these genes differs from country to country, whereas the genes encoding carbapenemases such as IMP, VIM, and NDM type metallo-β-lactamases have been found in all continents. Almost all types of transferable carbapenemases, except SIM- 1, have been detected in P. aeruginosa, and the prevalence of carbapenem-resistant isolates of P. aeruginosa is gradually increasing.
It is of concern that transferable plasmids carrying some of the resistance genes are mobile among a wide range of unrelated Gram-negative bacteria which increases the antimicrobial resistance transfer rate causing increasing treatment complications. Recent findings about antibiotic resistance have been even more concerning and warning. Liu et al. reported the first evidence of plasmid-mediated colistin resistance from China. Colistin (or polymyxin E) belongs to the family of polymyxins. The members of this class of antibiotics such as polymyxin B and colistin have been the last resort for antibiotic treatment of carbapenem-resistant bacteria such as P. aeruginosa isolates and Enterobacteriaceae. Resistance to polymyxins was previously reported to occur via chromosomal mutations; however, new evidence suggests plasmid-mediated resistance through the mobilization of the mcr-1 gene which consequently confers resistance to colistin. This gene was discovered in E. coli strain SHP45 collected from agricultural products. It is more concerning that the plasmid carrying mcr- 1 was mobilized into K. pneumoniae and P. aeruginosa via conjugation. This finding has triggered serious concerns about the emergence of pan-drug-resistant Gram-negative bacteria leading to almost untreatable infections. Recent findings provided some evidence of the spreading high-risk of clone ST654 of P. aeruginosa containing the genomic blaNDM−1 resistance gene which also conferred resistance to colistin. It is likely that blaNDM−1 was acquired via genetic exchange between P. aeruginosa and K. pneumoniae isolate in the same patient [17].
Adaptive Resistance Mechanisms
Compared to other types of resistance mechanisms, adaptive mechanisms are not really well understood. Adaptive resistance is an unstable and transient form of resistance, which is induced in the presence of specific antibiotics and other environmental stresses. This type of resistance mainly relies on induced alterations in gene expression and protein production or alterations in antibiotic targets and it is reversal upon removal of external stimuli leading to re-gaining susceptibility. This mechanism has been seen mediating the resistance of P. aeruginosa isolates to β-lactams, aminoglycosides, polymyxins and fluoroquinolones.
It has been seen that once strains encounters certain concentrations of antibiotics, they can tolerate higher concentrations in subsequent exposures, while cross-resistance to other antibiotics may occur as well. Furthermore, these alterations may link to other physiological events triggered by other stimuli and stresses as well as mutations in some specific genes.
Using isolates from CF patients, it was shown that adaptive resistance of P. aeruginosa to fluoroquinolones such as ciprofloxacin is due to multiple mutations in the knownresistance genes including the gyrA, gyrB, nfxB, and orfN which were concomitant with mutations in the genes involved in cyclic di-GMP signaling. Mutations of nfxB were prevalent leading to loss of function of NfxB transcriptional repressor and consequently leading to the overproduction of MexCD-OprJ efflux pump. This efflux pump is an important determinant of resistance to fluoroquinolone antibiotics. On the other hand, another study showed that expression of the mexCD-oprJ genes depends on the sigma factor AlgU and leads to resistance to the biocide chlorhexidine. AlgU is wellknown stress response sigma factor which positively regulates overproduction of alginate in mucoid isolates [18].
Another group showed that P. aeruginosa can acquire and lose resistance in the presence and absence of colistin, respectively. This occurred via adaptive multiple mutational mechanisms and genetic reversio. It was also demonstrated that resistance to certain polycationic antimicrobials such as aminoglycosides, polymyxins and cationic antimicrobial peptides can be mediated by altering the lipid A structure in LPS. This was caused by multiple mutations in cognate regulatory proteins such as the two-component systems PhoP-PhoQ, PmrA-PmrB, CprR-CprS, and ParR-ParS. Other studies showed that further and complex genetic alterations affecting regulatory pathways including those causing amino acid substitutions in these cognate regulatory proteins such as PhoQ and PmrB are involved in polymyxin resistance. This is why the mechanism of resistance of P. aeruginosa to colistin was found to vary among isolates. Interestingly, this study showed that the acquisition of colistin resistance via many amino acid substitutions is reversible in colistin-susceptible revertants. However, even in the absence of colistin, resistance was preserved for some time and emergence of revertants may not occur so fast.
QS-Dependent Antibiotic Resistance
Some direct and indirect evidences have been found linking the QS systems with antibiotic resistance mechanisms in P. aeruginosa, but further exploration is needed for better understanding. Using clinical strains of P. aeruginosa, it was shown that the las system positively links to the expression of mexY gene encoding the inner-membrane drug/H+ antiporter protein MexY which is a key subunit of the MexXY-oprM complex known as a major determinant of aminoglycoside resistance. On the other hand, some studies showed that CF-infecting strains with the common lasR loss-of-function mutations were more resistant to therapeutic antibiotics such as tobramycin, ciprofloxacin and ceftazidime. The reported antibiotic resistances in the lasR mutants were attributed to increased β-lactamase activity, bacterial metabolic adaptation or metabolic shifts. However, the relationship of antibiotics susceptibility with the rhl encoded QS system and production of C4-HSL signals remains unclear. Some supporting evidence was obtained by, treating P. aeruginosa biofilms with ciprofloxacin which upregulated the production and secretion of the virulent factor LasB, which is under the control of Rhl QS system [19].
Furthermore, two independent studies reported that the clinical strains of P. aeruginosa with QS-deficient phenotypes and negative for the production of QS-dependent virulence factors could cause infections and tend to be less susceptible to antimicrobial agents. However, it was not shown how these mechanisms might link to each other while many of these clinical strains could also form biofilms with antibiotic resistance traits and many regulatory pathways for biofilm development are under the control of QS systems. Zhao et al. reported some supporting information showing the importance of QS systems in both biofilm formation and antimicrobials induced expression of ampC. Earlier studies showed that by overexpressing the chromosomal type 1 β-lactamase, QSdependent virulence factors were reduced and strains were less virulent. Also, Kong et al. analyzed the dual role of the AmpR transcriptional regulator where it positively regulated β-lactamases and negatively regulated the virulence factors through QS systems.
Balasubramanian et al. analyzed co-regulatory and transcriptional networks of three co-existing mechanisms involved in β-lactam resistance, alginate production and modulation of virulence factor expression. They showed that while AmpR positively and negatively regulates β-lactamases and QS-dependent proteases, respectively, there is an intimate crosstalk between the AmpR regulon and the master regulator AlgU which positively regulates alginate production. This gave more insight into the complexity of such co-existing networks. Recent findings also showed that high levels of cyclic di-GMP mediated by the SagS regulator contributes to elevated antibiotic resistance via BrlR regulon-dependent upregulation of cognate genes encoding MexAB-OprM and MexEF-OprN multidrug efflux pumps [20].
The periplasmic TpbA tyrosine phosphatase was also reported as a regulatory candidate for linking QS signaling and biofilm formation. This protein was shown to be positively regulated by the las QS system at transcriptional level. Upon production of TpbA and its phosphatase activity in the periplasm, the cyclic di-GMP synthesizing protein TpbB is dephosphorylated at a tyrosine residue in periplasmic domain leading to inactivation of TpbB and a reduction in cyclic di-GMP levels and in turn Pel production, hence inhibiting biofilm development. TpbA-dependent cyclic di-GMP reduction was also linked to increasing eDNA release by cell lysis.
Overall, the reason for inconclusive information about the relation of the QS system and antibiotic resistance mechanisms is based on the fact that there are various layers of regulatory pathways associated with both QS systems and antibiotic resistance mechanisms in P. aeruginosa. Therefore, understanding the interplay between hierarchical QS systems and various antibiotic resistance mechanisms needs further exploration [21].
Adaptive Radiation for Persistence
Adaptation to the surrounding environments is an extraordinary capability of P. aeruginosa. It enables P. aeruginosa to inhabit diverse ecological niches such as colonization of various hosts as well as long term persisting infections. The adaptation process is designated as adaptive radiation by which initial clones would diversify into a variety of genotypes and phenotypes over time until the most favorable and adapted descendants are selected for long term persistence. A typical example is adaptation of P. aeruginosa isolates to the CF airways. Various studies have shown that initial colonization of CF lungs is caused by wild-type strains existing in the environment. For bacteria, the CF lungs encompass various stresses such as oxidative stresses and immune responses and inter-species competition followed by antibiotic treatment. Therefore, initial clones undergo substantial adaptation processes to survive such hostile environments [22].
Here, adaptive radiation is mainly due to intense genetic adaptations leading to the thousands of generations displaying diverse genotypes and phenotypes that emerge in vivo, while subjected to selection pressure imposed by the CF lung milieu. Therefore, selected variants display different genotypes when compared with initial wild-type colonizers and persist in the CF lungs leading to clonal expansion within patients and establishment of chronic infections. By assessing a wide selection of phenotypes, Workentine et al. showed that the overall population structure in one chronically infected patient can be much more heterogeneous in phenotypes than what has been previously documented. Furthermore, it has been reported that transmission of strains from patient to patient can result in the coexistence of highly divergent bacterial lineages.
Aim and Objectives
Aim
1. To evaluate the antibacterial resistance pattern in Pseudomonas aeruginosa;
2. To provide base for formulating rational antibacterial guidelines to treat the infections caused by pseudomonas aeruginosa.
Objectives
Pseudomonas aeruginosa is a Gram-negative and ubiquitous environmental bacterium. It is an opportunistic human pathogen capable of causing a wide array of life-threatening acute and chronic infections, particularly in patients with compromised immune defence. It has been of particular importance since it is the main cause of morbidity and mortality in cystic fibrosis (CF) patients and one of the leading nosocomial pathogens affecting hospitalized patients while being intrinsically resistant to a wide range of antibiotics.
Materials and Methods (M&M1)
In this study a total of 18 P.aeruginosa isolates were obtained out of 25 pus samples between a period of one year(Feb 2021 to April 2021). The isolates were selected on the basis of their growth characteristics on Blood agar, MacConkey agar and Nutrient agar medium. Colonies were subjected to battery of biochemical tests to identify species. Antimicrobial susceptibility testing of all confirmed P. aeruginosa isolates was performed by Kirby –Bauer disc diffusion method and results were interpreted according to CLSIs guidelines.
Samples collection and culture
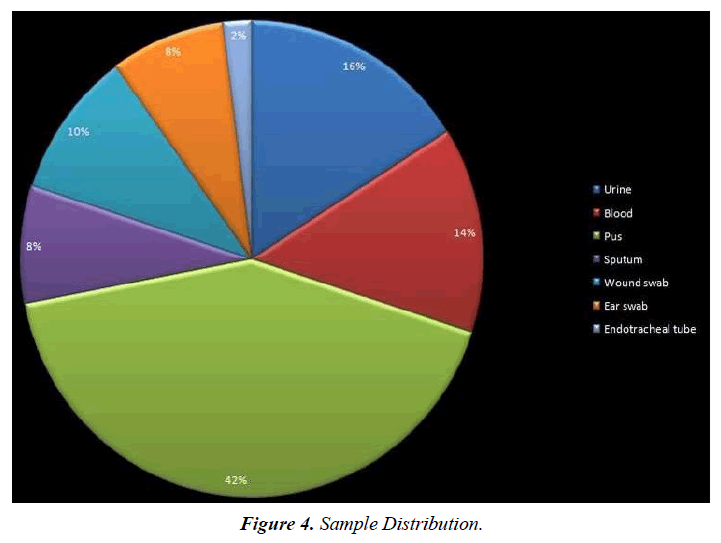
In the current cross-sectional study, a total of 3248 clinical samples including wound, urine, sputum, blood, feces, and trachea were collected from February 2021 to April 2021 from Ashwini hospital, Cuttack. Samples were cultured on selective media including MacConkey agar, and Blood agar, Cled agar (24 h on 37 °C) based on bacteriology standards (Figure 4).
Identification of Bacterial Isolates
Isolates were identified based on the Bergeys̓ microbiology book guidelines. In brief, conventional biochemical tests including oxidase, catalase, motility, metabolic procedure such as citrate, Indol production, Methyl red, Voges– Proskauer, and presence of lysine decarboxylase, and arginine dehydrogenase enzymes were performed.
Re-confirmation of P. aeruginosa isolates by PCR
Molecular identification of P. aeruginosa was performed by targeting the gene for P. aeruginosa and isolates using specifically designed primers which produced a 956 and 353 bp PCR product. Amplification was done based on the mentioned references with slight modification on the Gene Amp PCR system (Applied Biosystem, USA) in the total volume of 25 μl containing 14 μl master amplicon (Biolab, New England, UK), 1 pmol of each forward and reverse primers, a minor amount of colony as a template and 9 μl distilled water. The first cycle of denaturation was at 95 °C for 5 min, followed by 25 cycles at 94 °C for 1 min, then at 58 °C, (55 for blaoxa51) for 1 min, at 72 °C for 60 s, and finally a terminal extension for 5 min. PCR products were visualized by 1% agarose gel (KBC, Max Pure agarose, Spain) containing 0.5 μg/mL DNA Safe Stain dye in gel image analysis system (UVitec, Cambridge, UK). P. aeruginosa ATCC 27853 were used as the positive and negative control strains, respectively.
Antibiotic susceptibility tests
Isolates confirmed as P. aeruginosa and by biochemical and molecular tests underwent the disc diffusion susceptibility test based on the clinical and laboratory standards institute (CLSI, M100S 26th edition breakpoints) guidelines. In short, a 0.5 McFarland suspension of each isolate was inoculated on a whole plate surface Mueller–Hinton agar plate by streaking the swab in back and forth motions. Antimicrobial impregnated discs including amikacin (30 μg), ceftazidime (30 μg), cephalexin (30 μg), ciprofloxacin (5 μg), imipenem (10 μg), meropenem (10 μg), gentamycin (10 μg), tobramycin (10 μg), and cotrimoxazole (25 μg) were put on the surface of the agar, and the plates were incubated for 24 h at 37 °C. Following incubation, inhibition zone sizes to the nearest millimeter were measured using a ruler. Using published CLSI guidelines, susceptibility, or resistance of the organism to each tested drug was determined. Interpretation of Antibiotic susceptibility to evaluated MDR and XDR isolates was performed based on the european centre for disease prevention and control (ECDC) instructor as well [23].
The present study was conducted in the Department of Microbiology, Ashwini Hospital. All pus samples received from various departments from Feb. 2021 to march. 2021 were processed for isolation and identification of P. aeruginosa was made according to the Standard microbiological techniques. Blood agar, MacConkey agar and Nutrient agar were used as growth media for the culturing of samples. The plates were then incubated at 37°C for 24 hours to get the growth and were then processed further for identification using standard procedures. P.aeruginosa was identified by -Gram staining, motility test and biochemical tests like- oxidase test, O/F test, and growth at 420 C.8 Antibiotic sensitivity pattern of P. aeruginosa isolates to Gentamicin (10 mcg), Ciprofloxacin (5 mcg), Cefotaxime (30 mcg), Ceftazidime (30 mcg), Amikacin (30 mcg), Imipenem (10 mcg), Meropenem (10 mcg), Cefoperazone/ Sulbactum (75/30 mcg), Cefpirome (30 mcg), Aztreonam (50 mcg), Ceftazidime / Clavulanic acid (30/10 mcg), Piperacillin/Tazobactum (100/10 mcg), Piperacillin (100 mcg), Polymyxin (300 u), Colistin (10mcg) was investigated by Kirby-Bauer method on Mueller Hinton Agar (MHA). The final bacterium inoculation conc. was approx 108 cfu/ml that was equal to 0.5 McFarland. MHA plates were incubated overnight at 370 , and the diameter of each inhibition zone was measured with special scale supplied by Himedia Mumbai.
P. aeruginosa strains were isolated from pus samples received from various departments. Prevalence of P.aeruginosa was 22.41%.The highest isolation rate of P.aeruginosa was from surgery department, as shown in. The highest percentage of isolates was from males (71.9%) and of age group 41-50,61- 70 (21%) years each. Most of the isolates were found to be highly sensitive to Colistin (95.4%), Polymyxin B (95%), Levofloxacin (83.3%), Imipenam (70%), Netilmicin (66%) and Piperacillin + Tazobactum (64.5%) . However,they showed resistance towards Ofloxacin (65%), Piperacillin (64%), Ceftazidime (56.3%), Cefoprazone (58%), Cefipime (55%), Aztreonam (53%), Cefaprazone + sulbactum (46%) and Gentamicin (45%). As the bacterial strains that show resistance to three or more categories of antibiotics are defined as multidrug resistanct (MDR) strains, MDR strains of P.aeruginosa isolated in this study were 24%. Fourteen P. aeruginosa isolates (6 from department of ICU, 5 from department of Surgery, 2 from department of OBG and 1 from department of Emergency) were totally resistant to Cephalosporins, Aminogycoside, Fluoroquinolones and Carbapenems, showing Multidrug resistance (MDR) [24]. P.aeruginosa presents a serious therapeutic challenge for treatment of both community acquired and nosocomial infections. Infections caused by P.aeruginosa are notoriously difficult to treat due to its intrinsic ability to resist many classes of antibiotics as well as its ability to acquire resistance. Our study measures the rate of isolation of P.aeruginosa is 22.44%. The occurrence of P.aeruginosa is found to be higher in males, inpatients in age group >60,41 years and in surgery department reported that most of isolates were found to be highly sensitive to Colistin (95.4%), Polymyxin B (95%), Levofloxacin (83.3%), Imipenem (70%), Netilmicin (66%) and Piperacillin + Tazobactum (64.5%), Sensitivity pattern of P.aeruginosa nearly coincides. P.aeruginosa showed resistance towards Ofloxacin (65%), Piperacillin (64%), Ceftazidime (56.3%), Cefoprazone (58%), Cefipime (55%), Aztreonam (53%), Cefaprazone + sulbactum (46%) and Gentamycin (45%), which was comparable w
References
- World Health Organization. Antimicrobial resistance: global report on surveillance. World Health Organization; 2014.
- Dereli N, Ozayar E, Degerli S, et al. Three-year evaluation of nosocomial infection rates of the ICU. Rev Bras Anestesiol. 2013;63:79-84.
- Fraimow HS, Tsigrelis C. Antimicrobial resistance in the intensive care unit: mechanisms, epidemiology, and management of specific resistant pathogens. Crit Care Clin. 2011;27(1):163-205.
- Micek ST, Wunderink RG, Kollef MH, et al. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: impact of multidrug resistance. Crit Care. 2015;19:1-8.
- http://www.whocc.no/atc_ddd_index/
- Spilker T, Coenye T, Vandamme P, et al. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42(5):2074-9.
- AmirMoezi H, Javadpour S, Golestani F. Identification of different species of Acinetobacter Strains, and determination of their antibiotic resistance pattern and MIC of Carbapenems by E-Test. Hormozgan Med J. 2016;20(1):45–51.
- Patel J, Weinstein M, Eliopoulos G, et al. M100 Performance standards for antimicrobial susceptibility testing. United State: Clinical and Laboratory Standards Institute. 2017:240.
- El-Solh AA, Hattemer A, Hauser AR, et al. Clinical outcomes of type III Pseudomonas aeruginosa bacteremia. Crit Care Clin. 2012;40(4):1157.
- Hauser AR. Pseudomonas aeruginosa virulence and antimicrobial resistance: two sides of the same coin?. Crit Care Clin. 2014;42(1).
- Reboud E, Elsen S, Bouillot S, et al. Phenotype and toxicity of the recently discovered exlA‐positive Pseudomonas aeruginosa strains collected worldwide. Environ Microbiol. 2016;18(10):3425-39.
- Wood TL, Wood TK. The HigB/HigA toxin/antitoxin system of Pseudomonas aeruginosa influences the virulence factors pyochelin, pyocyanin, and biofilm formation. Microbiologyopen. 2016;5(3):499-511.
- Giamarellos-Bourboulis EJ, Koussoulas V, Panagou C, et al. Experimental sepsis using Pseudomonas aeruginosa: The significance of multi-drug resistance. Int J Antimicrob Agents. 2004;24(4):357-61.
- Giamarellos-Bourboulis EJ, Tzepi I, Tsovolou I, et al. Impact of multidrug resistance on experimental empyema by Pseudomonas aeruginosa. Respiration. 2011;82(1):46-53.
- Giamarellos-Bourboulis EJ, Plachouras D, Tzivra A, et al. Stimulation of innate immunity by susceptible and multidrug-resistant Pseudomonas aeruginosa: An in vitro and in vivo study. Clin Exp Immunol. 2004;135(2):240-6.
- Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-81.
- Giakkoupi P, Vourli S, Polemis M, et al. Supplementation of growth media with Zn2+ facilitates detection of VIM-2-producing Pseudomonas aeruginosa. J Clin Microbiol. 2008;46(4):1568.
- Alvarez‐Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol. 2007;65(1):153-65.
- Amato SM, Fazen CH, Henry TC, et al. The role of metabolism in bacterial persistence. Front Microbiol. 2014;5:70.
- Amato SM, Orman MA, Brynildsen MP. Metabolic control of persister formation in Escherichia coli. Mol Cell. 2013;50(4):475-87.
- Amiel E, Lovewell RR, O'Toole GA, et al. Pseudomonas aeruginosa evasion of phagocytosis is mediated by loss of swimming motility and is independent of flagellum expression. Infect Immun. 2010;78(7):2937-45.
- Avrain L, Mertens P, Van Bambeke F. RND efflux pumps in P. aeruginosa: An underestimated resistance mechanism. Antibiotic Susceptibility. 2013;26321:26-8.
- Behera B, Vishnu GA, Chatterjee S, et al. Emerging technologies for antibiotic susceptibility testing. Biosens Bioelectron. 2019;142:111552.
- Ayers M, Sampaleanu LM, Tammam S, et al. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. J Mol Biol. 2009;394(1):128-42.
- Bains M, Fernández L, Hancock RE. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ. 2012;78(18):6762-8.
- Bajolet-Laudinat O, Girod-de Bentzmann S, Tournier JM, et al. Cytotoxicity of Pseudomonas aeruginosa internal lectin PA-I to respiratory epithelial cells in primary culture. Infect Immun. 1994;62(10):4481-7.
- Balasubramanian D, Kong KF, Jayawardena SR, et al. Co-regulation of β-lactam resistance, alginate production and quorum sensing in Pseudomonas aeruginosa. J Med Microbiol. 2011;60(Pt 2):147.
- Balasubramanian D, Kumari H, Jaric M, et al. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res. 2013;42(2):979-98.
- Balasubramanian D, Kumari H, Mathee K. Pseudomonas aeruginosa AmpR: An acute–chronic switch regulator. Pathog Dis. 2015;73(2):1.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref