Research Article - Biomedical Research (2017) Volume 28, Issue 21
Rab8b regulates kidney epithelial cell migration and lamellipodia formation
Hanyu Zhu1,2#, Minghui Wu1,2#, Dong Zhang1,2*, Wenjia Geng1,2*, Qiuxia Han3, Qingyi Wang4, Xiaoli Yang1,2, Yan Shi3, Guangyan Cai1,2 and Xiangmei Chen1,2
1Department of Nephrology, Chinese PLA General Hospital, Chinese PLA Institute of Nephrology, Beijing, PR China
2State Key Laboratory of Kidney Diseases, National Clinical Research Center of Kidney Diseases, Beijing Key Laboratory of Kidney Disease, Beijing, PR China
3Department of Nephrology, the First Affiliated Hospital of Zhengzhou University, 1 East Jianshe Road, Zhengzhou, PR China
4Center for Computational and Integrative Biology, Massachusetts General Hospital, Harvard Medical School, Boston, MA 02114, USA
#These authors have equally contributed to this work.
- *Corresponding Author:
- Dong Zhang
Department of Nephrology
Chinese PLA General Hospital
Chinese PLA Institute of Nephrology
Beijing, PR China
Wenjia Geng
State Key Laboratory of Kidney Diseases
National Clinical Research Center of Kidney Diseases
Beijing Key Laboratory of Kidney Diseases
Beijing, PR China
Accepted on September 06, 2017
Abstract
Cell migration is a central process by which multicellular organisms can be properly established and maintained. Lamellipodia are an actual motor that is critical for cell migration. Rab8b, a member of Rab small GTPase, is known to play a role in secretion, endocytosis and ciliogenesis. However, it remains unknown whether Rab8b is involved in cell migration. In the present study, we show that Rab8b is ubiquitously expressed in the mouse tissues including kidney. We further show that depletion of Rab8b in the kidney epithelial cell line mIMCD-3 resulted in reduced velocity of cell migration, which is likely associated with defective formation of lamellipodia. Our data suggest Rab8b plays an important role in cell migration and lamellipodia formation.
Keywords
Rab8b, Rab8a, Knockdown, Cell migration, Lamellipodia.
Introduction
Cell migration is the process by which cells move from one location to another on certain substratum through distinct motility modes, such as mesenchymal, amoeboid or collective migration. This cell activity is a fundamental event to establish and maintain proper organization of multicellular organisms [1]. The collective cell migration is a hallmark of the tissue remodeling events that underlie embryonic morphogenesis, wound repair and cancer invasion [2]. Failed or defective cell migration can result in many defects during development or life-threatening abnormalities, such as immunosuppresion, autoimmune diseases, defective wound repair, or tumor dissemination [3,4]. Lamellipodia are actin cytoskeletal projections on the leading edge of the cells and they are an actual motor which pulls the cell forward during cell migration [5,6]. Lamellipodia formation has long been known to be actin-dependent and controls directed cell migration [7].
The Rab family of small GTPases plays important roles in regulating vesicular transport in all eukaryotes. In the past decade, a line of evidences have linked some members of the Rab family, such as Rab5, Rab8, Rab21 and Rab25, to the mechanistic aspects of cancer cell migration and invasiveness [8]. Rab8b was originally cloned from rat basophilic leukaemia cell line, RBL.2H3, and encodes a 207-aa protein Rab8b which is highly homologous (82%) to Rab8a with distinct C-terminal domain [9]. Rab8b is mapped on chromosome 9 and spans around 76 kb of genomic DNA that contains 8 exons and 7 introns. Northern blot analysis revealed that Rab8b is ubiquitously expressed in different tissues of rat with highest levels of expression of Rab8b in the spleen, testis and brain while Rab8a exhibited a different expression pattern [9]. The first interacting partner of Rab8b, termed TRIP8b, was identified a few years later, which might define a role of Rab8b in the regulated secretary pathway in AtT20 cells [10]. Otoferlin, another interacting partner of Rab8b, is defective in a human autosomal recessive deafness form, suggesting that Rab8b may be also involved in this disease [11]. Recently, human RAB8B was identified to be an essential evolutionary conserved component of Wnt/β-catenin signaling through the regulation of LRP6 activity and endocytosis [12]. Rab8b together with Rab8a is essential for several apical transport pathways but insufficient for ciliogenesis [13]. More recently, it was reported that Rab8b, like Rab7 and Rab24, has a key role in autophagosome maturation [14]. Rab8b is also specifically involved in the trafficking of West Nile Virus (WNV) particles from recycling endosomes to the plasma membrane [15]. Rab8b was found to be associated with the actin, intermediate filament, and microtubule cytoskeletal networks, and participates in adherens junction dynamics in the testis [16]. Although Rab8a, the isoform of Rab8b, was shown to be involved in cancer cell migration and invasiveness, it remains unknown whether Rab8b itself has a similar role in this cellular process.
In the present study, we investigated the role of Rab8b in cell migration and lamellipodia formation in a kidney epithelial cell line. We also examined the expression profile of Rab8b in mouse tissues.
Material and Methods
Western blot
Total protein was extracted from 1 month-old mouse tissues. Tissue lysates were denatured with 6 × sample buffer and boiled at 100°C for 10 min. The samples were then loaded into 12% SDS-PAGE. After gel running, proteins were electrotransferred onto a nitrocellulose membrane (Bio-Rad), followed by blocking with 5% non-fat milk in TBST (1 × TBS with 0.05% Tween-20) for 1 h at room temperature (RT). The membrane was then blotted with primary antibody, Rab8b (Proteintech, Cat. No. 11792-1-AP) or GAPDH, for 2 h at RT. The membrane was then washed with TBST for 10 min/3 times followed by incubation with the secondary antibody (goat anti-mouse or goat anti-rabbit) for 1 h at RT. After washing with TBST for 10 min/3 times, membrane-bound antibody was detected with ECL (enhanced chemiluminescence) kit.
Knockdown experiments
Four Rab8b shRNAs (Rab8b KD1~KD4) cloned in pLKO.1 vector were purchased from Sigma. The target sequences are as follows: Rab8b KD1 (clone ID: MmSH00276827): ccggccggtctaagaagaccagtttctcga (located in exon 8; within ORF); Rab8b KD2 (clone ID: MmSH00276839): ccggcaaatgtgatatgaacgacaactcga (located in exon 5; within ORF); Rab8b KD3 (clone ID: MmSH00277633): ccggccgaacaattacgacagcatactcga (located in exon 3; within ORF); Rab8b KD4 (clone ID: MmSH00277641): ccgggccaagaactaacagaactttctcga (located in exon 8; within 3’ UTR). The Rab8b shRNAs, packaging plasmid pCMV-dR8.91 and envelope plasmid VSV-G were co-transfected into HEK293 cells with lipofectamine 2000 to produce shRNA-bearing lentiviruses. 18 h after transfection, the DMEM media were replaced with media containing 30% FBS. The HEK293 cells were cultured for an additional 24 h.
The viruses produced in the media were then collected to infect mIMCD-3 cells (80~90% confluence) with additional fresh DMEM/F12 media containing polybrene. Puromycin selection (2 μg/ml) was started at ~48 h post-transduction. Based on the knockdown efficiency, Rab8b KD4 cells at earlier passage were used for later experiments.
Wound healing assay
The Rab8b knockdown cells and scramble control cells were cultured with DMEM/F12 media in 6-well plate until reaching confluent. The wound was created along the diameter of the well with a 20 μl tip. The cells were washed with DPBS twice and fresh DMEM/F12 media were replenished. Images were taken along the wound at the same spots at different time points, including 0 h, 6 h and 9 h. Prior to imaging at each time point, the cells were washed with DPBS to remove the floating or dead cells.
Immunofluorescence staining
Cells were grown on 22 mm coverslips (Warner Instruments), placed in 6-well plates (Corning), and cultured for 1-2 d until the cells reached appropriate confluence. The cells were fixed with 4% FPA for 15 min at RT and then washed twice with PBS, permeabilized with 0.3% Triton X-100 for 5 min. The cells were washed twice more with PBS, and then blocked with 5% BSA (dissolved in PBS) for 1 h at RT. After blocking, the cells were incubated with Rab8b antibody (Proteintech, Cat. No. 11792-1-AP) or rhodamine phalloidin (Invitrogen, Cat. No. R415) overnight at 4°C. The next day the cells were washed with PBS 5 min twice, and then incubated with Alexa Fluor 488 (Green; Invitrogen, Cat. No. A11001) and/or Alexa Fluor 594 (Red; Invitrogen, Cat. No. A11012) conjugated secondary antibodies for 1 h at RT. The cells were then washed twice with PBS again, and mounted with Prolong Gold Antifade with DAPI (Invitrogen, Cat. No. P36935). Images were taken by using SPOT software with a Nikon TE1000 microscope system.
Result
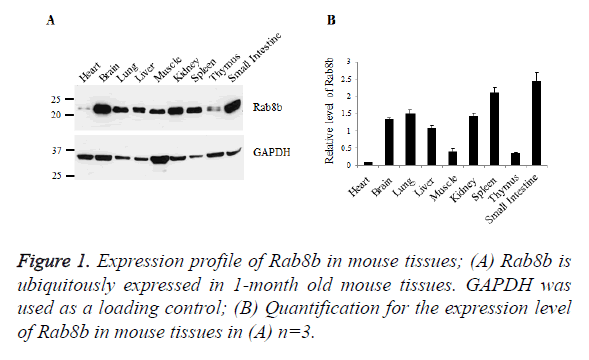
Rab8b is ubiquitously expressed in mouse tissues
To examine the expression profile of Rab8b in mice, we collected samples from mouse heart, brain, lung, liver, muscle, kidney, spleen, thymus and small intestine. Tissue lysates were blotted with specific antibody of Rab8b. The data showed that Rab8b is ubiquitously expressed in mouse tissues with variable levels. In the brain, lung, liver, kidney, spleen and small intestine, Rab8b is highly expressed, while in the heart, muscle and thymus, Rab8b has much less abundance, especially in the heart (Figures 1A and 1B).
Knockdown of Rab8b in the kidney epithelial cells
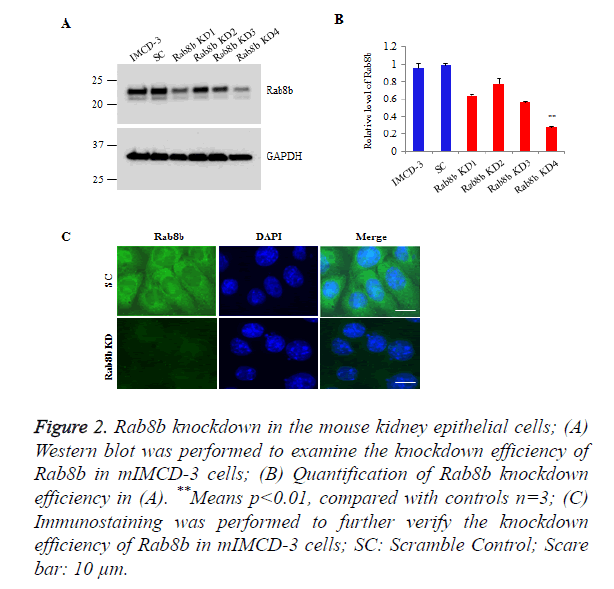
To investigate the function of Rab8b in the kidney, we knocked down Rab8b in a mouse inner medullary collecting duct cells (mIMCD-3), a commercially well-characterized mouse kidney cell line. The cells were transduced with lentiviruses bearing shRNAs which are targeted to distinct sites of Rab8b transcript and then stably selected with puromycin. The Western blot data show that Rab8b KD4 has the highest knockdown efficiency, compared with other 3 targets (Figures 2A and 2B). We further performed immunostaining to confirm the knockdown efficiency for Rab8b KD 4. The data show that Rab8b in highly depleted in the knockdown cell line compared with the scramble control cells (Figure 2C). These data suggest that the Rab8b-depleted cell line was successfully established.
Figure 2. Rab8b knockdown in the mouse kidney epithelial cells;
(A) Western blot was performed to examine the knockdown efficiency of Rab8b in mIMCD-3 cells;
(B) Quantification of Rab8b knockdown efficiency in (A). **Means p<0.01, compared with controls n=3;
(C) Immunostaining was performed to further verify the knockdown efficiency of Rab8b in mIMCD-3 cells; SC: Scramble Control; Scare bar: 10 μm.
Rab8b regulates cell migration in the kidney epithelial cells
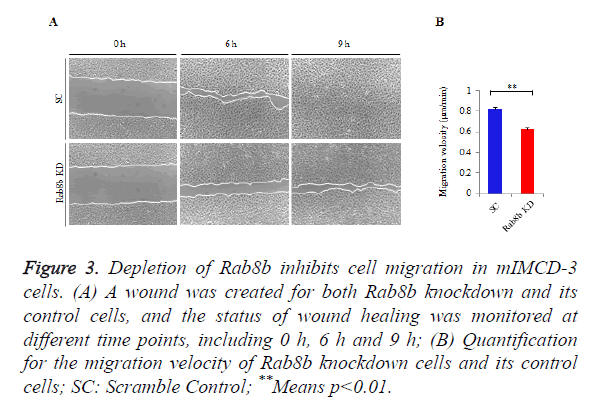
Rab8b is known to be involved in multiple cellular processes, while it remains unknown whether it is involved in cell migration. To address this question, we performed wound healing assay in the Rab8b knockdown cell line.
A similar size of cell wound was created in the Rab8b knockdown and its control cells. The velocity of wound healing was monitored at three different time points, including 0 h, 6 h and 9 h.
The data showed that cell migration was much slower in the Rab8b knockdown cells compared with the control cells (Figure 3A). The velocity of cell migration in the control cells was about 0.8 μm/min, while it was reduced to be 0.6 μm/min in the Rab8b knockdown cells (Figure 3B).
Figure 3. Depletion of Rab8b inhibits cell migration in mIMCD-3 cells.
(A) A wound was created for both Rab8b knockdown and its control cells, and the status of wound healing was monitored at different time points, including 0 h, 6 h and 9 h;
(B) Quantification for the migration velocity of Rab8b knockdown cells and its control cells; SC: Scramble Control; **Means p<0.01.
Rab8b regulates lamellipodia formation in the kidney epithelial cells
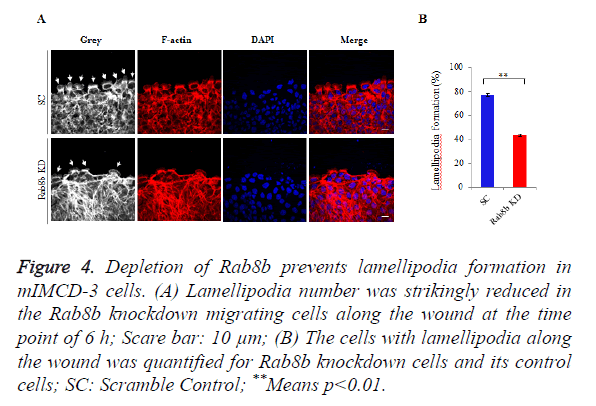
Because lamellipodia formation is a key determinant of cell migration, we went to examine the lamellipodia number in the migrating cells within the created wounds. We counted the number of lamellipodia and the total number of cells along the wound. The data showed that lamellipodia number was largely reduced in the Rab8b knockdown cells compared with the control cells (Figure 4A). The percentage of cells with lamellipodia was significantly lower in the Rab8b knockdown cells (Figure 4B).
Figure 4. Depletion of Rab8b prevents lamellipodia formation in mIMCD-3 cells.
(A) Lamellipodia number was strikingly reduced in the Rab8b knockdown migrating cells along the wound at the time point of 6 h; Scare bar: 10 μm;
(B) The cells with lamellipodia along the wound was quantified for Rab8b knockdown cells and its control cells; SC: Scramble Control; **Means p<0.01.
Discussion
Rab8b was first discovered in the rat basophilic leukaemia cell line and it is ubiquitously expressed in all types of animal tissues. The transcript of Rab8b was shown to be more abundant in the heart, brain, spleen, liver, skeletal muscle and testis of the rats while it was less in the lung and kidney [9]. Another study from different group showed that Rab8b can be detected in the brain, testis, heart, kidney and spleen of the rat with highest level in the brain and lowest level in the heart [16]. In our study, we found that at the protein level Rab8b is more abundant in the brain, lung, kidney and spleen of the 1- month old mouse. The discrepancy is likely resulted from the difference in species, animal age and/or post-transcriptional modification.
The homologous protein of Rab8b, Rab8a, was previously shown to promote the formation of new surface extensions through reorganization of actin and microtubules, and play an important role in directed membrane transport to cell surfaces in fibroblasts [17]. This process is likely monitored by a Rab8- specific activator Rabin8 [18]. Rab8a was also shown to play a key role in membrane traffic during neuronal process outgrowth and identified as a critical component of the cellular machinery that specifically drives the local delivery of AMPA-type glutamatergic receptors (AMPARs) into synapses [19,20]. By a yeast two-hybrid assays and Fluorescent Resonant Energy Transfer (FRET) techniques, Rab8a, but not Rab8b, was identified to be an interacting partner of myosin Vb, which is important in recycling a variety of receptors [21]. Rab8a is known to be involved in regulating the actin-dependent movement of melanosomes [22]. In line with this finding, scientists also found that Rab8a regulates the formation of actin containing structures such as filopodia, lamellipodia, protrusions and primary cilia, and is known as one of the important regulators for cell migration, epithelial polarization and ciliogenesis [23]. Based on this line of evidences, we hypothesized that Rab8b might also have a function in regulating the formation of lamellipodia and cell migration since it is an isoform of Rab8a. We knocked down Rab8b in a kidney epithelia cell line, and examined the velocity of cell migration and the capability of lamellipodia formation. The data from our present study strongly support our hypothesis. Our findings are important advance for the understanding of Rab8b in addition to its roles in endocytosis, ciliogenesis and autophagosome maturation [12-14].
It is known that Rho family proteins are pivotal regulators of actin and adhesion organization and control the formation of lamellipodia. Of the Rho family proteins, Rac, Cdc42 and RhoG are required for the protrusion of lamellipodia [4]. Therefore, our future study might be directed to the connection between Rab8b and Rho signaling pathways.
Acknowledgement
This study was supported by the National Key R & D Program of China (2016YFC1305502), the National Natural Science Foundation of China (No. 61471399; No. 61671479) and the Innovation Nursery Fund of PLA General Hospital (No. 15KMZ04).
Disclosure of Interest
The authors declare that they have no conflicts of interest.
References
- Trepat X, Chen Z, Jacobson K. Cell migration. Comprehensive Physiol 2012; 2: 2369-2392.
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature Rev Mol Cell Biol 2009; 10: 445-457.
- Franz CM, Jones GE, Ridley AJ. Cell migration in development and disease. Developmental Cell 2002; 2: 153-158.
- Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Sci 2003; 302: 1704-1709.
- Small JV, Stradal T, Vignal E, Rottner K. The lamellipodium: where motility begins. Trends Cell Biol 2002; 12: 112-120.
- Machesky LM. Lamellipodia and filopodia in metastasis and invasion. FEBS Lett 2008; 582: 2102-2111.
- Ballestrem C, Wehrle-Haller B, Hinz B, Imhof BA. Actin-dependent lamellipodia formation and microtubule-dependent tail retraction control-directed cell migration. Mol Biol Cell 2000; 11: 2999-3012.
- Tang BL, Ng EL. Rabs and cancer cell motility. Cell Motility Cytoskeleton 2009; 66: 365-370.
- Armstrong J, Thompson N, Squire JH, Smith J, Hayes B, Solari R. Identification of a novel member of the Rab8 family from the rat basophilic leukaemia cell line, RBL.2H3. J Cell Sci 1996; 109: 1265-1274.
- Chen S, Liang MC, Chia JN, Ngsee JK, Ting AE. Rab8b and its interacting partner TRIP8b are involved in regulated secretion in AtT20 cells. J Biol Chem 2001; 276: 13209-13216.
- Heidrych P, Zimmermann U, Bress A, Pusch CM, Ruth P, Pfister M, Knipper M, Blin N. Rab8b GTPase, a protein transport regulator, is an interacting partner of otoferlin, defective in a human autosomal recessive deafness form. Hum Mol Genet 2008; 17: 3814-3821.
- Demir K, Kirsch N, Beretta CA, Erdmann G, Ingelfinger D, Moro E, Argenton F, Carl M, Niehrs C, Boutros M. RAB8B is required for activity and caveolar endocytosis of LRP6. Cell Rep. 2013; 4: 1224-1234.
- Sato T, Iwano T, Kunii M, Matsuda S, Mizuguchi R, Jung Y, Hagiwara H, Yoshihara Y, Yuzaki M, Harada R, Harada A. Rab8a and Rab8b are essential for several apical transport pathways but insufficient for ciliogenesis. J Cell Sci 2014; 127: 422-431.
- Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ 2014; 21: 348-358.
- Kobayashi S, Suzuki T, Kawaguchi A, Phongphaew W, Yoshii K, Iwano T, Harada A, Kariwa H, Orba Y, Sawa H. Rab8b Regulates Transport of West Nile Virus Particles from Recycling Endosomes. J Biol Chem 2016; 291: 6559-6568.
- Lau AS, Mruk DD. Rab8B GTPase and junction dynamics in the testis. Endocrinol 2003; 144: 1549-1563.
- Peranen J, Auvinen P, Virta H, Wepf R, Simons K. Rab8 promotes polarized membrane transport through reorganization of actin and microtubules in fibroblasts. J Cell Biol 1996; 135: 153-167.
- Hattula K, Furuhjelm J, Arffman A, Peranen J. A Rab8-specific GDP/GTP exchange factor is involved in actin remodeling and polarized membrane transport. Mol Biol Cell 2002; 13: 3268-3280.
- Huber LA, Dupree P, Dotti CG. A deficiency of the small GTPase rab8 inhibits membrane traffic in developing neurons. Mol Cell Biol 1995; 15: 918-924.
- Gerges NZ, Backos DS, Esteban JA. Local control of AMPA receptor trafficking at the postsynaptic terminal by a small GTPase of the Rab family. J Biol Chem 2004; 279: 43870-43878.
- Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell 2007; 18: 2828-2837.
- Chabrillat ML, Wilhelm C, Wasmeier C, Sviderskaya EV, Louvard D, Coudrier E. Rab8 regulates the actin-based movement of melanosomes. Mol Biol Cell 2005; 16: 1640-1650.
- Peranen J. Rab8 GTPase as a regulator of cell shape. Cytoskeleton (Hoboken) 2011; 68: 527-539.