Review Article - Journal of Genetics and Molecular Biology (2023) Volume 7, Issue 1
Olfactory Receptors in Mammals including Bats
R. Steffi Christiane, T. Karuppudurai*Department of Animal Behaviour & Physiology, School of Biological Sciences, Madurai Kamaraj University, Madurai, India
- *Corresponding Author:
- T Karuppudurai
Department of Animal Behaviour & Physiology
School of Biological Sciences
Madurai Kamaraj University, Madurai, India
E-mail: tkdurai@gmail.com
Received: 15-Dec-2022, Manuscript No. AAGMB-22-83068; Editor assigned: 17-Dec-2022, PreQC No. AAGMB-22-83068 (PQ); Reviewed: 31-Dec-2022, QC No. AAGMB-22-83068; Revised: 03-Jan-2023, Manuscript No. AAGMB-22-83068 (R); Published: 10-Jan-2023, DOI:10.35841/aagmb-7.1.131
Citation: Karuppudurai T. Olfactory receptors in mammals including bats. J Genet Mol Biol. 2023;7(1):131
Abstract
Among mammals, olfaction is employed to varied degrees in all facets of life, including the detection of food, the avoidance of predators, and social interaction. Additionally, different species have different olfactory capacities. Dogs and rodents, for example, depend on smell to travel, forage, and communicate. This is reflected in the number of functional OR genes present in these species. The mammalian genome contains the biggest gene family, the olfactory receptor (OR) gene repertoire, which encompasses over 1,000 functional OR genes, each of which codes for a distinct OR and is expressed sequentially in the cells of the olfactory epithelium. Binding of odors to ORs executes odor perception, which starts a signaling cascade to the brain's olfactory bulb through a G-protein coupled receptor. 13 monophyletic groups are supported by phylogenetic analysis of the nucleotide sequences of mammalian OR genes. Despite being highly annotated in the completed human and mouse genomes and making up 3 to 6% of mammalian genes, we still do not fully understand which odorants bind to which receptors and how this intricate process translates into perceiving a certain smell. Olfactory receptor genes expression can be discerned by looking at reasonably closely related species that have a wide variety of diets. Bats are potentially useful for this purpose. Bats (Chiroptera) represent one of the most fascinating mammal groups for studying OR gene expression studies. Fruit bats mainly employ olfaction in food detection and recently, many studies have revealed that the olfaction is linked to dietary specialization. Nevertheless, across very short distances, smell cues from mammals like bats can be more effective than vocalization. Conversely, several studies have examined the significance of olfaction in bat food acquisition and detection. While numerous comparative studies of nectarand fruit-eating bats have examined how olfaction and foraging ecology are related. In India, in fact, until recently, there is no systematic study of OR gene expression pattern in mega and micro bats. We employ molecular biology and bioinformatics techniques to identify the unique and diverse OR genomic repertoire in bats. Our preliminary results suggest that the total number of OR genes and families vary widely among both fruit and insect eating bats. If reflected in the diversity of OR genes, the large range of sensory specializations and modalities in bats could be used to explain the variety and uniqueness of the bat OR repertoire. In this review, the general structure and function of mammals' olfactory receptors, including those found in bats, are summarized.
Keywords
Chiroptera, Bats, Olfaction, G-protein Coupled Receptor, OR Genes
Introduction
Olfaction is a vital sense that helps living individuals to get chemical information from their surroundings. The olfactory system processes two types of stimuli: (a) general odorants, which are small molecules derived from food or the environment that indicate the presence of food, fire, or predators, and (b) pheromones, which are molecules released by members of the same species and convey social or sexual cues [1]. Chemosensory receptors are classed as odorant or pheromone receptors based on the ligands that activate them. Sensory neurons in the periphery that express either odorant or pheromone receptors convey signals to discrete odor- and pheromone-processing areas in the brain, eliciting different behavioural and neuroendocrine responses. Pheromones engage narrowly tailored receptors that activate sexually dimorphic neuronal circuits in the brain, whereas general odorants activate receptors in a combinatorial way [1].
Olfaction is one of the most important types of sensory perception in mammals, and it is the foundation for the exceptional sensitivity necessary to differentiate environmental and sexual signals. As a result, olfactory receptor (OR) genes are the largest gene superfamily, accounting for 6% of proteincoding genes in a typical mammalian genome (total OR genes/ total number of protein-coding genes in dog: 1100/19,300). Olfaction is employed to variable degrees in many areas of life among vertebrates, including food detection, predator avoidance, and social communication [2,3]. The olfactory receptor (OR) gene family is the biggest in the mammalian genome, with over 1,000 functional OR genes, each of which codes for a single OR and is expressed in the olfactory epithelium cells in a sequential order [3-5]. The binding of smells to ORs causes a signaling cascade to occur in the olfactory bulb of the brain, which is mediated by a G-protein coupled receptor. Dogs and rats rely on scent to travel, forage, and communicate, but humans rely more on visual and aural signals [6]. Most animals rely on their sense of smell to survive. It is used to discover food, avoid threats, identify partners and offspring, and mark territory. Olfactory receptors (ORs) expressed in the olfactory epithelium of the nasal cavity detect various odour molecules in the environment [7,8].
Olfactory System
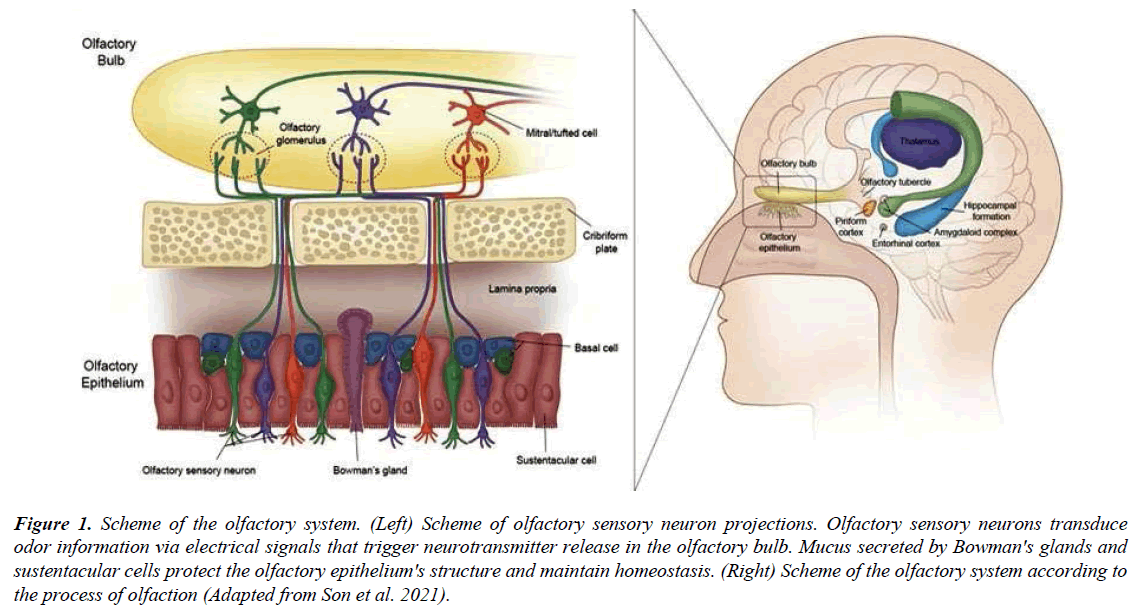
To monitor the external environmental chemical changes from the brain, the olfactory system in the nose acts as a window (Figure 1). This olfactory system is made up of four subsystems: i) the main olfactory epithelium (MOE), ii) the vomeronasal organ (VNO), iii) the septal organ (SO) of masera and iv) the Grueneberg ganglion (GG) [9]. All olfactory systems have four characteristics in common. They are: 1) the presence of odorant binding proteins in the fluid overlying the receptor cell dendrite; 2) the requirement of G protein-coupled receptors as odorant receptors (even though some sensory neurons, such as in Caenorhabditis elegans and mammals, may use transmembrane guanylate cyclase receptors); 3) the use of a two-step signalling cascade in odorant transduction; and 4) the presence of functional structures of the first central target in the olfactory pathway [10] (Figure 1). All of these traits may reflect independently evolved adaptations that provide us with vital information about how the nervous system interprets odorant inputs [11].
Figure 1: Scheme of the olfactory system. (Left) Scheme of olfactory sensory neuron projections. Olfactory sensory neurons transduce odor information via electrical signals that trigger neurotransmitter release in the olfactory bulb. Mucus secreted by Bowman's glands and sustentacular cells protect the olfactory epithelium's structure and maintain homeostasis. (Right) Scheme of the olfactory system according to the process of olfaction (Adapted from Son et al. 2021).
Olfactory Perception
The olfactory perception begins when the odorants interact with the highly specific biological machinery i.e., ORs present in the nasal/olfactory epithelium (OE) [12-14]. Each odorant produces a unique pattern of neuronal signal that consists of signal intensity, time and quality of odorant stimuli. This further stimulates a specific population of olfactory sensory neuron (OSNs) present in Olfactory Epithelium which process and transduce the signals at neurological level [14,15]. Neural signal perception produces a representation known as "smell" which is represented by various perpetual descriptors such as fruity, woody, rose, etc. Since odors are insubstantial, have a complex molecular basis and are perceived individually, the process of olfaction is challenging [14,16,17]. Odors linked with any material (flower, plant, etc.) are made up of a variety of odorants, some of which play a major role and others which play a little role. Due to the fact that one odorant can have several scents (eugenol methyl ether has 27 odour perceptions), two structurally distinct compounds can have nearly the same olfactory profile (cis-3-hexenol, nonadienal, ligustral exhibits green odor). Carvone enantiomers, (R)-(- )-carvone (spearmint odour) and (S)-(+)-carvone (caraway odour) have unique scents due to a tiny structural change; this intricate connection was mostly unknown until now [14].
Olfactory Receptor (OR) Genes
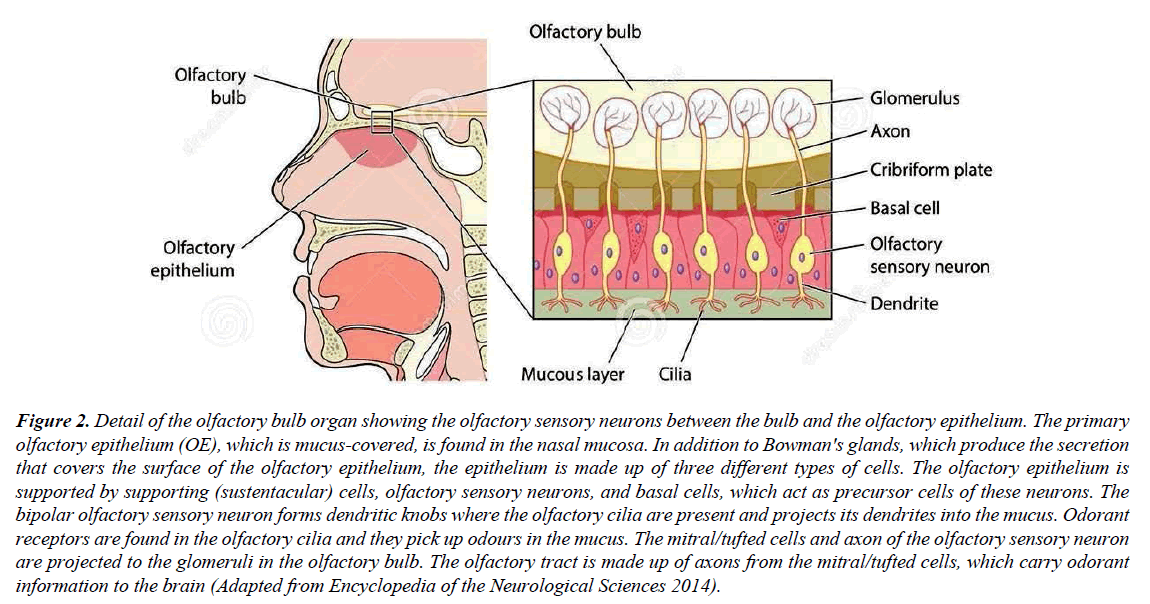
Chemical signals, often odorants stimulate ORs in the cilia of olfactory sensory neurons (OSNs) which is located in the olfactory epithelium (OE). This stimulation of ORs by the odorants, contribute to the sense of smell in the brain Figure 2; [18]. The OR gene family is the largest among G protein-coupled receptors (GPCRs) with over 1,000 genes on the mouse chromosome and over 450 genes in the human genome [9,19]. ORs in mammals belong to the rhodopsinlike family of G protein-coupled receptors (GPCRs) with a 7-transmembrane domain helix bundle arrangement that share several sequence motifs having a number of highly conserved amino acids in the transmembrane domains (TMs). These motifs are also present in mouse ORs (MORs) along with more than a few OR-specific motifs [19,20]. Since the overall sequence identity is low (25%) among rhodopsin-like GPCRs, there is scope for significant deviations in the ligand binding pockets and in inter-helical contacts, which makes the study of GPCR ligand binding sites more challenging [20]. Among species, the total number of OR genes differ broadly. Remarkably, the largest repertoire of intact OR genes ever identified within a single species was in African elephants having 2230 OR pseudogenes. This outnumbered the largest previously identified repertoire in rats [21].
Figure 2: Detail of the olfactory bulb organ showing the olfactory sensory neurons between the bulb and the olfactory epithelium. The primary olfactory epithelium (OE), which is mucus-covered, is found in the nasal mucosa. In addition to Bowman's glands, which produce the secretion that covers the surface of the olfactory epithelium, the epithelium is made up of three different types of cells. The olfactory epithelium is supported by supporting (sustentacular) cells, olfactory sensory neurons, and basal cells, which act as precursor cells of these neurons. The bipolar olfactory sensory neuron forms dendritic knobs where the olfactory cilia are present and projects its dendrites into the mucus. Odorant receptors are found in the olfactory cilia and they pick up odours in the mucus. The mitral/tufted cells and axon of the olfactory sensory neuron are projected to the glomeruli in the olfactory bulb. The olfactory tract is made up of axons from the mitral/tufted cells, which carry odorant information to the brain (Adapted from Encyclopedia of the Neurological Sciences 2014).
Olfactory Receptor Gene Structure and Organization
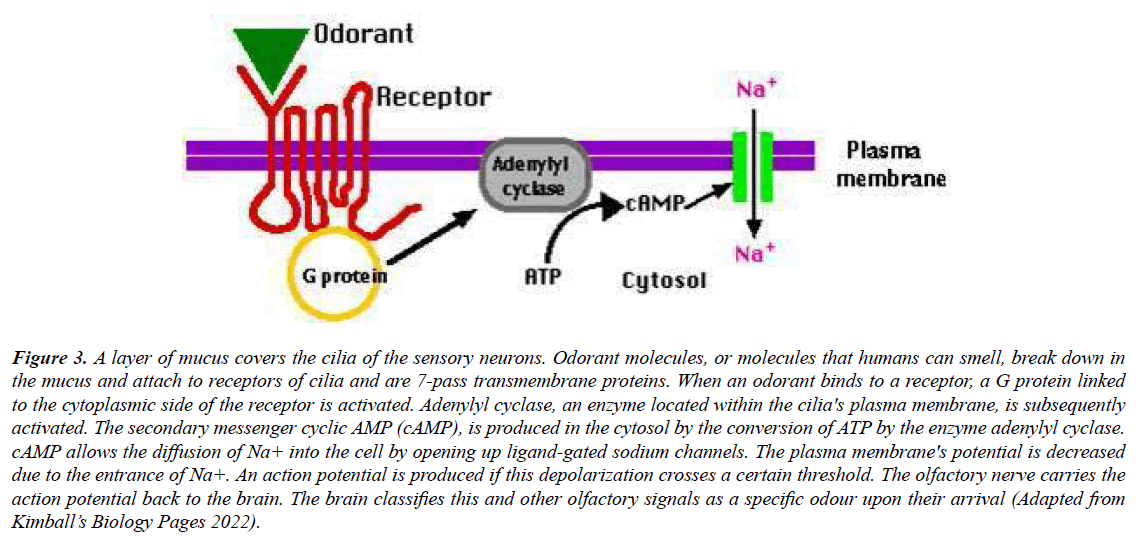
The structure of OR genes are unusual with an intron less coding region, short up and downstream non-coding exons as well as corresponding introns. OR genes form very condensed units as the transcription start site is situated on one end while the polyadenylation signal is on the other end. This transcription starts site and the polyadenylation signal are located in close proximity (1-10 kb) to the coding sequence (Figure 3). This organization favors the evolutionary dynamics of this gene family. Different isoforms of OR mRNAs which form the same protein are present as the upstream exons of several OR genes were found to be alternatively spliced [18,22,23].
Figure 3: A layer of mucus covers the cilia of the sensory neurons. Odorant molecules, or molecules that humans can smell, break down in the mucus and attach to receptors of cilia and are 7-pass transmembrane proteins. When an odorant binds to a receptor, a G protein linked to the cytoplasmic side of the receptor is activated. Adenylyl cyclase, an enzyme located within the cilia's plasma membrane, is subsequently activated. The secondary messenger cyclic AMP (cAMP), is produced in the cytosol by the conversion of ATP by the enzyme adenylyl cyclase. cAMP allows the diffusion of Na+ into the cell by opening up ligand-gated sodium channels. The plasma membrane's potential is decreased due to the entrance of Na+. An action potential is produced if this depolarization crosses a certain threshold. The olfactory nerve carries the action potential back to the brain. The brain classifies this and other olfactory signals as a specific odour upon their arrival (Adapted from Kimball’s Biology Pages 2022).
In the mammalian genome, OR genes are widely dispersed and are found on all chromosomes. OR genes are extensively distributed throughout mammalian genomes and may be found on almost every chromosome. They are usually found in a variety of places, each with a different number of genes. Non-OR interspersed genes are not found in OR clusters in general [24]. Depending on the number of added repetitive sequences, intergenic distances range from less than 5kb to more than 50kb. Several clusters have since been studied in depth indicating that each of them may contain members of several subfamilies or even families, implying that OR cluster [19,22]. Alternatively, genes from the same subfamily may appear in many clusters, implying that clusters were partially or totally duplicated. Various groups of interspersed repeating elements account for a large share of cluster sequences. These repetitions are thought to be involved in the many transposition/duplication events that occur in the OR repertoire during evolution [18].
Classification of Olfactory Receptors
OR sequences are classified into two classes based on the evolutionary data: Class I and Class II. Class I receptors were proposed to be specialized in identifying the water-soluble odorants since they have earmarks of the family initially found in aquatic animals [25, 26, 27]. Only terrestrial animals express class II receptors. The OR repertoire can be divided into Class I receptors (binds water-borne odorants) and Class II receptors (binds mainly volatile odorants). These classes are further fragmented into four families (OR 51, OR52, OR55, and OR56) and nine OR gene families (OR 1/3/7, OR 2/13, OR 4, OR 5/8/9, OR 6, OR 10, OR 11, OR 12, and OR 14) respectively, with each family also having a range of smaller subfamilies [28].
A comparison of the structural properties of both receptor classes from different species indicated that they differ mostly in the sequence of the second extracellular loop, which was proposed to play a role in ligand selectivity [25]. Class I genes in humans are clustered on chromosome 1, while class II genes are present on all chromosomes except chromosome 20 and Y. Class I ORs have a pseudogene proportion of 52 percent, while class II ORs have a pseudogene fraction of 77 percent [26,27]. Frog (Xenopus laevis) has two types of ORs: one that is comparable to fish ORs (class I) and another that is similar to mammalian ORs (class II). Most of ORs in mammals are classified as class II, however, even class I ORs do express in mammals [19,25,29]. In humans and mice, for example, there are over 100 class I ORs; unexpectedly, a high proportion of these are potentially functional implying that some ancient ORs have been preserved and may even perform a particular role in mammals [18,33].
Olfactory Receptors (OR) Nomenclature
The Olfactory Receptors (OR) sequences have been classified using a variety of nomenclatures. OR genes have been classified into families and subfamilies by Glusman and Lancet, with members of a particular family sharing a protein sequence identity of >40% (PID) and subfamily members sharing a PID of >60% [26,30]. According to this classification, the human genome has 17 families, four of which have more than 100 members. Based on chromosomal location and phylogenetic research, a study suggested a new nomenclature [30]. A new nomenclature approach for mouse OR sequences have been proposed that integrates phylogenic links and protein identity. Despite the fact that databases seek to present the correspondences of a particular OR in many nomenclatures, the issue remains perplexing and should be clarified in the near future [19,30].
Evolution of Olfactory Receptor Genes
In macrosmatic animals such as dogs and mice, the number of OR sequences (functional and nonfunctional genes) contained in the genome ranges from roughly 1,500 to about 800 in microsmatic primates. Humans (387) and platypus (262) have a modest repertoire of functional OR genes with rat (1,284) and mouse (1,194) having the greatest [18,31,32]. Many OR genes have been added and lost during mammalian evolution. The high turnover of OR genes in vertebrates is likely due to the functional need for various olfactory skills in different evolutionary lineages. The marsupial lineage saw the most gene family increase, with at least 750 new genes. Similarly, the rodent lineage has accumulated around 400 genes. The number of genes lost in the primate lineage, on the other hand, is substantially higher than in other lineages [4,18].
Structural Features of Odorant Receptor Proteins
OR proteins have seven hydrophobic, putative membranespanning domains, which are common to all GPCRs. GPCRs are divided into three groups according on their main sequence: A, B, and C. ORs, like rhodopsin, belong to GPCR class A, according to this categorization, because of their domain structure [18,34]. OR proteins are around 320-25 amino acid residues long on average, with variations in length due to varying N- and C-terminal regions. A well conserved NXS/T consensus for N-linked glycosylation may be found in the N-terminal region that is accessible extracellularly [18]. Several conserved amino acid motifs distinguish ORs from other GPCRs, including an LHTPMY motif within the first intracellular loop, the most distinctive MAYDRYVAIC motif at the end of transmembrane (TM) domain 3 (TM3), a very short SY motif at the end of TM5, an FSTCSSH stretch at the beginning of TM6, and PMLNPF in TM7. Despite the fact that these sequences change slightly between species, they have been utilized to identify OR genes in a variety of genomes. More than 80 short motifs have been found through extensive comparative analysis [18,32,35], some of which are distinctive for different subfamilies or species and have been linked to ligand binding. Seven cysteine residues are highly conserved, with two of them considered to have a role in the protein's structural stability. Two of these (at locations 97 and 179) are found in all GPCRs and are thought to create a disulfide connection between extracellular loops 1 and 2, whereas the other five are found only in ORs [18].
Olfactory Signal Transduction
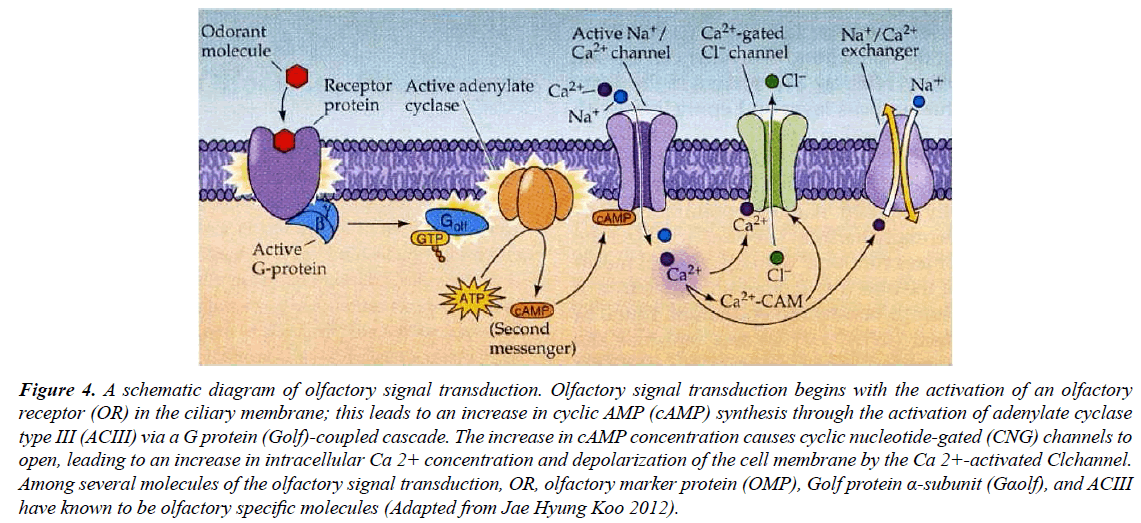
Olfactory receptor cells transduce the odour signal by linking it to one or more downstream effector molecules in addition to expressing GPCRs. As previously mentioned, heteromeric GTP binding proteins and intracellular second messengers are used by GPCRs to couple to downstream effectors (Figure 4). Given the odorant's relatively brief dwell time on the receptor, recent data point to an exceedingly modest elementary response, i.e., a low probability that the ligandbound receptor would even activate one G protein molecule [36]. The majority of olfactory second messengers target ion channels, which when opened change the membrane potential of the cell and cause a graded, voltage-dependent response that results in all-or-nothing electrical impulses (action potentials, often called "spikes"). The graded shift in membrane potential has an impact on how frequently action potentials go to the central nervous system [37] (Figure 4). Olfactory neurons use phosphoinositide-derived signals and cyclic nucleotides as two key intracellular signaling pathways. There is no obvious evolutionary tendency in the usage of one signaling cascade over the other, and these pathways appear to be active in a wide variety of animals. The cyclic nucleotide signaling in vertebrate olfactory receptor neurons is the best understood [38]. The olfactory cyclic nucleotide-gated ion channel, whose activation permits calcium entrance into the cell and then activates a calcium triggered chloride current in a two-step activation cascade, is the target of cyclic nucleotide signaling in these cells. The excitatory receptor potential is largely produced by the later current [37,39].
Figure 4: A schematic diagram of olfactory signal transduction. Olfactory signal transduction begins with the activation of an olfactory receptor (OR) in the ciliary membrane; this leads to an increase in cyclic AMP (cAMP) synthesis through the activation of adenylate cyclase type III (ACIII) via a G protein (Golf)-coupled cascade. The increase in cAMP concentration causes cyclic nucleotide-gated (CNG) channels to open, leading to an increase in intracellular Ca 2+ concentration and depolarization of the cell membrane by the Ca 2+-activated Clchannel. Among several molecules of the olfactory signal transduction, OR, olfactory marker protein (OMP), Golf protein α-subunit (Gαolf), and ACIII have known to be olfactory specific molecules (Adapted from Jae Hyung Koo 2012).
In nematodes (Caenorhabditis elegans) and arthropods (lobster) cyclic nucleotide signaling also seems to be involved in olfactory transduction [40,41]. Then cyclic nucleotide signaling, the role of phosphoinositide signaling plays a crucial role in the activation of crustacean olfactory receptor cells. There, a calcium-sensitive putative lobster homolog of the TRP family of ion channels serves as the target of phosphoinositide signaling [42]. The channel can be targeted directly by 3-phosphoinositides or indirectly by gating extracellular calcium from an associated plasma membrane InsP3 receptor when odorants activate both the PLC- and PI3Kmediated arms of this signaling cascade [37,43] (Figure 4).
Other, phylogenetically varied animals, such as worms, insects, fish and mammals have also been linked to phosphoinositide signaling in some capacity There is presumptive requirement for the receptor cell to use various signaling cascades to encode the magnitude of receptor binding, hence it is unknown whether individual olfactory receptor cells use both cyclic nucleotide and phosphoinositide signaling cascades [44,45]. However, several signaling cascades may enable the cell to integrate responses to complex odorants when coupled to various receptors or to various places on the same receptor in a ligand-specific way, with potentially significant ramifications for odour coding. The use of both intracellular signaling pathways by olfactory receptor cells in arthropods lobster, rat and mammals suggests that signaling through these pathways may play a fundamental role in olfactory transduction, though this role needs further extensive study [37,40,45].
Odorants
Chemicals that bind to olfactory receptors and are transduced into electrical signals are called odorants. In terms of its volatility, each odorant has different attributes [46]. These attributes include how the odorant distributes in the environment, dissolves in liquids or a carrier gas and adheres to the surfaces. These attributes can change depending on certain environmental conditions such as humidity, pressure, temperature or even the characteristics of a container [46]. Odorant mixtures elicit less variable and faster responses than pure odorants. In terms of volatility, each odorant has diverse features, how it distributes in the environment, adheres to
surfaces, or dissolves in liquids or a carrier gas, and these features can alter depending on environmental conditions such as temperature, pressure, humidity, or even characteristics of the container [46]. When many odorants must be considered, the complexity of the problem becomes more challenging, both in terms of interactions between chemical components in the environment and interactions with receptors, because of its physical qualities, each odorant is unique [47-49]. Most of the time, odorants are part of a turbulent environment, and they produce very complex odor plumes. Indeed, both the physical features of the airflow and the odorants influence the spatiotemporal organization of odor plumes. Turbulence is determined by the parameters of the flow, while the interplay between diffusive and advective motion is determined by the qualities of the odorants. The "odor-landscape" describes the distribution of odorant concentration in space, with its valleys, crests, and plateaus [50,51].
Expression of Vertebrate OR genes
A specific vertebrate OR gene is expressed in a small subset of OSNs in the olfactory epithelium, which are interspersed with OSNs expressing other OR genes. Only one OR is expressed per OSN, according to single cell RT-PCR (reverse transcriptase-polymerase chain reaction) data and other techniques [52,53] Furthermore, even though each gene is represented by two alleles, a single OSN not only expresses an exclusive OR gene, but it also appears that only one allele is expressed in each particular neuron. Allelic exclusion describes the expression pattern in which OSNs express either the maternal or paternal allele in roughly equal numbers in the olfactory epithelium [30]. Furthermore, the epithelium of the mouse is separated into four zones, with each OR gene expressed in only one of them. Each zone covers about a quarter of the epithelium's surface [30,52] and each OR allele is expressed in a tiny proportion of neurons in an apparently random distribution within each zone, implying that the final selection phase is stochastic. The topographies of these zones in rodents are complicated, but they are commonly oriented anterior-posterior as stripes along distinct sections of the turbinates. This spatial structure in humans has yet to be validated, and its biological importance is unknown [30,54]. OR expression has also been found in the testis, which is an adult tissue. Approximately 10% of mammalian ORs are transcribed during male germ cell development and in mature spermatozoa. ORs have been found to be transcribed in both the olfactory epithelium and the testis [30,55,56]. ORs expressed in testes appear to be more conserved than ORs expressed in OSNs, at least in some domains. The physiological relevance of OR expression in the testis is uncertain, however it is thought to have a role in sperm chemotaxis to the oocyte or sperm maturation [30].
Ligand Specificity of Olfactory Receptors
Individual olfactory sensory neurons react to a wide range of odorants, and each cell has its own agonist potency order, demonstrating that olfactory neurons are very diversified and extensively tuned. Based on the assumption that each olfactory sensory cell expresses just one OR subtype, it appears that olfactory receptors have a rather non-specific ligand range. Elucidating the ligand/receptor specificity, like with other orphan receptors, necessitates determining the responsiveness of a different receptor type, which is generally accomplished by expression in heterologous cell types and high-throughput screening tests [57,58]. OR genes have been shown to be extremely difficult to produce in heterologous systems, owing to a lack of appropriate receptor protein folding or membrane targeting. Some of these issues were overcome by approaches that used either a homologous in vivo expression system transfected with recombinant adenovirus and assessed by electrophysiological recordings or engineered OR chimeric receptors with the N-terminal "membrane-import-sequence" of either rhodopsin or serotonin receptors in heterologous cells monitored by imaging approaches [58-60]. The olfactory system computes information from combinations involving any of around a thousand receptor types, similar to how the visual system employs three receptor types (three opsin-subtypes of the three cone populations) to make sense of all perceivable colours. The system's ability to encode an infinite number of scents is explained by the various potential combinations. Instead of dedicating a single odour receptor to a single odour, the olfactory system employs a "alphabet" of receptors to produce a specific odour response; in this view, a different receptor type plays a role in encoding very different odours in the same way that a different letter plays a role in forming very different words [58].
Combinatorial Odor Coding
The olfactory system is generally believed to use "combinatorial coding" In this model, one odorant may be recognised by several ORs and one OR may be able to recognise a variety of odorants [61]. Instead, each OR does not have a one-to-one association with an odorant. In the end, various odorants are modelled as various combinations of activated ORs. For more than 15 years, researchers have been working hard to find ligands for ORs [62,63]. A study was conducted which tested 93 odorants against 464 ORs and effectively deorphanized 10 human and 52 mouse ORs [64]. Their findings demonstrate the validity of the combinatorial coding system. They also showed that certain ORs are "specialist" ORs that are narrowly tuned and only bind to a small number of structurally related odorants, whilst others are "generalist" ORs that are broadly tuned and bind to a large variety of odorants. However, the majority of ORs are still orphans, and little is known about how ORs and odorants interact [21]. Combinatorial coding states that odorants with nearly identical structures are recognised by different but overlapping sets of receptors, which explains why even minor changes in an odorant's structure can result in a dramatic shift in its perceived odour [61]. The odour of octanol changes from orange to rotten when the hydroxyl group is replaced by a carboxy group to form octanoic acid. It might also explain why an odorant's perceived quality changes as its concentration changes; for example, indole has a rotten odour when concentrated but is regarded as flowery when diluted [58]. The processes of odorant coding are currently poorly understood. However, both spatial and temporal patterns of activated neurons in olfactory centers are thought to contribute to odorant combinatorial coding [61,65].
Olfaction in Mammals
Previous studies surveyed the olfactory receptor (OR) repertoire encoded in 13 mammalian species and discovered that African elephants have the most characterised OR genes of any mammal, more than five times as many as dogs and more than twice as many as humans [21]. The authors examined genome sequences of 13 placental animals and discovered over 10,000 OR genes in order to describe the olfactory abilities of various mammals. Only 3 OR genes were shared and evolutionarily conserved across all 13 mammals, making each species' repertoire of OR genes very distinct. Unexpectedly, the African elephant, with approximately 2,000 OR genes, had the most extensive olfactory repertoire. Researchers targeted to comprehend the degree of heterogeneity in evolutionary dynamics between distinct OR genes and to identify the causes of such variation [8,21]. The OR gene repertoires in 13 species of placental mammals whose deep-coverage genome data are available for these purposes were thoroughly mapped. Among these 13 species, OGGs were identified using a phylogeny-based method, and ultimately, comparisons were made between OGGs [66]. These results showed that the evolutionary fates of OGGs differed significantly, and that this variation was related to the OR class, the degree of functional restrictions, the ligand selectivity, and the OR expression [21].
Recently, scientists have used molecular phylogenomic techniques to map the fate of species-specific gene duplication in 94 diverse mammalian taxa, in order to identify the OR gene families driving adaptation to various ecological niches. More than 70,000 OR gene sequences extracted from the entire genome were used for this analysis [28]. Results suggest that the presence of a functional vomeronasal organ is connected with statistically significant patterns of OR speciesspecific gene duplications for the first time. It was discovered that a novel association exists between the dietary niche of herbivory and a significant number of duplications in OR family 5/8/9. These findings also point to distinctions between sociable and solitary niches, suggesting that living alone may lead to a higher OR repertoire expansion. An essential way for new, diverse genotypes and phenotypes to emerge is through mechanisms like tandem gene duplication, segmental duplication, or whole-genome duplication [28,67,68].
African elephants may use olfaction to discriminate between members of their family, while Asian elephants can distinguish between enantiomer odorant pairs, making the elephant the mammal with the broadest range of ORs [7,21,69]. This remarkable olfactory ability is reflected in the African elephant's genome's detectable species-specific duplications (SSD) and the following putatively functional ORs. These results indicate a close relationship between the OR repertoire and the VNO, with a functional VNO being related to a rapid growth of the OR gene repertoire. Regarding OR gene development through SSD, no remarkable variations between rhythmic activity stages were found. Three times as many OR genes are produced through gene duplication in terrestrial mammals as in volant or aquatic mammals, which is indicative of the great variety of distinct terrestrial ecological niches to which mammals have successfully adapted. This work highlights the use of species-specific duplications in understanding the evolution of gene families by demonstrating how the OR repertoire has changed in relation to a range of ecological adaptations in mammals [28].
Researchers compared the functional OR sub genome repertoire across 50 phylogenetically and ecologically diverse mammals to study if the evolution of the OR gene repertoire has been influenced by habitat, sensory specialization, and other ecological traits; to clarify if there is a signature of OR natural selection within mammals; and to find which gene families are important in each ecological niche [7]. Principal component analysis (PCA) and Bayesian assignment tests were used to visualize and identify significant differences in the functional OR gene repertoire between aquatic, semi-aquatic, terrestrial, and flying/volant mammals, and to distinguish which OR families, if any, were driving these differences [7]. Despite the reported large disparities in the amount of pseudogenes among these taxa, ancestral state reconstructions show that the majority of Afrotherians, primates, and rodents appear to have preserved the ancestral mammalian distribution of functional OR genes [6]. Results suggests that more comparative and population genomic investigations of OR genes in ecologically varied taxa can help identify the genetic mechanisms underlying the development of sensory perception. In order to determine how many OR genes were most likely amplifiable in each species, the Gazey and Staley algorithm was used to compare laboratory-generated data with whole-genome sequence data for Myotis lucifugus. When the functional repertoire was compared as a whole, no significant difference was observed between low-coverage genomic data and laboratory generated data [7]. The recent neutralist theories [6] about the development of the olfactory subgenome was disproved and demonstrated that adaptive evolution significantly influences the make-up of the biggest gene family in the genome. These findings [70] demonstrate how the mammalian olfactory system has evolved to diverse habitats long hypothesized but never demonstrated before and reveal that various OR gene families are significant in various ecological niches [7].
Previous investigations suggest that a significant portion of the OR repertoire from seven phylogenetically and ecologically different mammalian species was amplified and sequenced using a combination of traditional laboratorybased polymerase chain reaction (PCR) and Next Generation Sequencing Technology (NGST) approaches. Even when available, reference genomes were not employed in the assembly procedure in order to retain a de novo assembly framework [71]. Comparing the existing information on OR genes from animals with fully sequenced genomes demonstrates that, although not amplifying the entire OR repertoire, the distribution of OR subfamilies in the data is consistent with chromosomal distributions. By using combinatorial approach, scientists suggest that NGST has revolutionised population genomics, deep level molecular phylogenetics, and evolutionary studies [72,73]. Despite significant improvements in sequencing speed and falling costs, the bioinformatics analysis and assembly of such massive data sets are the bottleneck in using this data. Modern biology must improve by creating algorithms and pipelines to more effectively filter and handle these data. A target-gene specific Illumina sequencing technique was created to better annotate and analyse these data, which can be used to any multigene family or species [71,72,74].
Olfaction in Bats
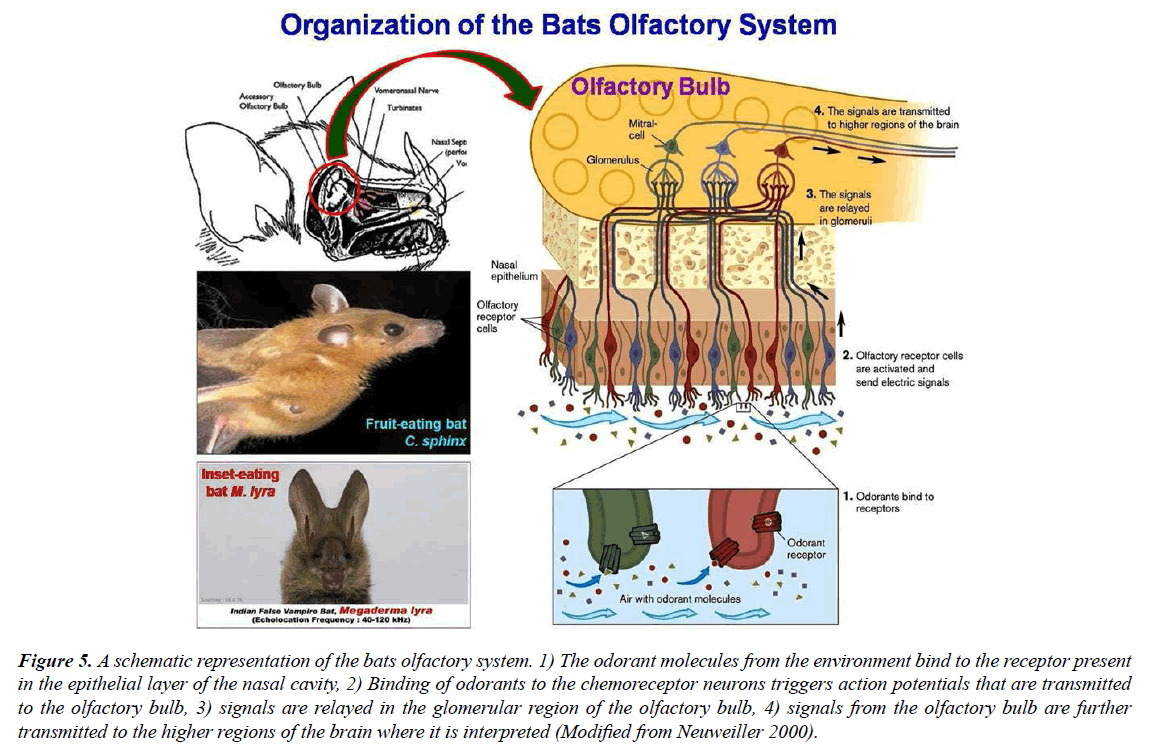
Bats are one of the most varied orders of mammals, second only to the mammalian Order Rodentia [75,76] with over 1,331 species divided into two suborders, the Megachiroptera (commonly known as Old World fruit bats) and the Microchiroptera [77]. Except for the Arctic and Antarctic, bats have been found around the world, and they make up more than 20% of all mammalian species [76]. Bats are essential for seed distribution, plant pollination, and the structure of forest ecosystems [78]. The bat olfactory system provides an excellent foundation for research on the distinctive and varied OR genetic repertoire. Bats ORs are a large family of genes that produce proteins that have direct interactions with chemical signals in the environment (Figure 5). In contrast to all other investigated mammals, bats had an extremely varied OR gene repertoire. With approximately 1,100 species (20% of the extant mammalian diversity) and a wide variety of niches, sensory modes, and dietary specializations [77,79], the chiropteran radiation provides an opportunity to investigate the ecological drivers of this unique and diverse OR genomic repertoire. Fruit bats (family Pteropodidae) are nonlaryngeal echolocators that use vision and olfaction instead of sound to orient themselves [80]. If reflected in the diversity of OR genes, the large range of sensory specializations and modalities in bats could be used to explain the variety and uniqueness of the bat OR repertoire. The frugivorous and nectarivorous bats' foraging strategies deeply rely on odor cues. Olfaction plays a crucial role in fruit bats in detecting the presence of ripe fruit (Figure 5).
Figure 5: A schematic representation of the bats olfactory system. 1) The odorant molecules from the environment bind to the receptor present in the epithelial layer of the nasal cavity, 2) Binding of odorants to the chemoreceptor neurons triggers action potentials that are transmitted to the olfactory bulb, 3) signals are relayed in the glomerular region of the olfactory bulb, 4) signals from the olfactory bulb are further transmitted to the higher regions of the brain where it is interpreted (Modified from Neuweiller 2000).
Previous studies suggest that genome size reduction in bats can be incompletely attributed to shortened introns and intergenic regions, which is a trend that is seen in birds [81]. This genome contraction might be an adaptation for powered flight and its associated high metabolic rates. A complete picture of gene and loss at a genome scale in bats is poorly understood though a large amount of genomic data has been generated for bats over the past four years [82-84]). Hence, scientists investigated the patterns of gene family evolution in bats and evaluated the average rate of gene gain and loss using a comparative genomics technique covering 20 mammalian genomes. Analysis was done to look whether this rate of gene gain and loss differs from that of other closely related lineages. Also, the families that underwent rapid evolution in the last common ancestor of both echolocating and non-echolocating bats, as well as the last common ancestor of bats was tried to analyze. Finally, whether the extremely small genome sizes of bats are related to rates of change in gene family size was discussed [84]. Results demonstrated that the protein-coding gene turnover in bat genomes is extremely flexible, but the pace of gene turnover seems to be comparable to that of their closely related Laurasiatherians. A potential trade-off between olfaction and other senses in auditory specialists is suggested by a high tendency of gene loss in ORs in bats, and echolocating lineages in particular. This trend was the opposite of that seen in both the non-echolocating bats and the carnivores investigated. A variety of suggested tree topologies for the links between laurasiatherian lineages seems to have little effect on these findings [84].
Neotropical fruit- and nectar-eating bats are known to use olfactory cues while foraging and are extremely sensitive to some fruit-typical odours [85,86]. These bats use echolocation for navigation. Fruit bats may detect and follow scent concentration gradients, especially when crawling, according to previous experimental study [87,88], but little is known about how their olfactory search tactics can be different from or enhance echolocation-based searches. The Jamaican fruit-eating bat (Artibeus jamaicensis) consumes a range of fruits, including bananas [89,90], and has been shown to have preferences for fruit odours when foraging [91-93]. In contrast, the limitations of flight may require bats to rely on other techniques, such as serial sampling or route-following, whereby bats are motivated by the presence of an alluring odour but need to sample each site in order to find the source of the odour [88,94,95].
Studies were conducted in a gregarious bat species Tylonycteris pachypus, to explore the idea that chemical discrimination is used to identify the mechanism involved in mother-pup and groupmate recognition. The findings suggested that in the mother pup odorant choice experiment, the mother bats can recognize their own pups by scent as expected. Feeding an alien offspring which is commonly known as allo-sucking was avoided by the mother and also the mother protects the pups from being killed by the predators. This is due to the effective recognition of pups by the mother [96]. In colonial mammals, including seal, seal lion, mouse and bat, mother– offspring olfactory recognition has been widely investigated and is known to be ubiquitous [97-99]. In the groupmate odorant choice (GOC) experiment, except for bats in group of male and female bats (female bat with scent samples from a female bat and a male bat of the same group), bats in the other group were more attracted to the scent from their groupmates regardless of sex. This finding is consistent with a previous study which shows that bats often exhibit a group scent profile that can be recognized by other groupmates [100]. No difference in preferences to the scent was observed from the same or opposite sex. With these results, the study concluded that the mother-pup and groupmate recognition of T. pachypus is accomplished by olfactory signals [101].
The link between external nasal morphology and potential olfactory tracking behaviour was evaluated in bat models. According to this study, the bat species that depend on olfaction for foraging had narrower nostrils when compare to other species that primarily depend on echolocation or hearing for foraging. This study states that it is not certain whether any ecological factors could be involved in diversity of bat nasal morphology [102]. All families of insectivorous bats had wider nostrils, while nectar feeding bat species had narrowest nostrils. Similar pattern was observed within Phyllostomidae family where insect eating bat species had wider nostril separation. Suitable odorant signals are required to stimulate foraging even during the absence of other food related signals, in several species of plant visiting Phyllostomid bats [86,87,103]. In the field where more captures recorded in odor-baited mist nets, bats were attracted to odor lure. But in the open fields, increased activity was observed around fruit odor lures [104]. In detection of ripe fruits or flowers over long distances, olfactory signals play an important role. However, bats could rely more on spatial memory to locate potential food resources and then rely on olfactory cues for fine-scale localization and discrimination [102,105].
Behavioural response of two fruit eating bats, Artibeus lituratus and Carollia perspicillata was evaluated in captivity to observe their olfactory preferences. Four experimental set up were established; Piper fruit vs Ficus fruit, piper oil vs ficus oil, piper oil vs ficus fruit and ficus oil with piper fruit. It was observed that Artibeus lituratus preferred ficus even though two out of four experimental set up were not statistically significant. This study indicates that the Ficus sp. were most common diet of Artibeus sp. [106-108]. Similar pattern was observed in A. lituratus, which chose Ficus fruits even with the increased supply of Piper (septum with essential oil extracted from approximately 300 g of fruit). It was identified that bat species preferred this Moraceae family due to high fibre content with low nitrogen and lipid concentration. Bats feed on large quantities of ficus to compensate this low nutrition content and supplement their diet with other species [109,110].
Earlier studies suggested that C. perspicillata showed high attempts on fruit and essential oil of Piper hispidum, even when the offer of ficus was greater (Piper fruit x Ficus oil tests). When compared to Moraceae genus, piper provides only a few mature fruits per night [110]. Co-evolution was observed between the Phyllostomid bats, with relationship between these bats and Piperaceae being well documented [111-113]. Studies suggest that the main intention of these bats is to feed on fruits which are rich in protein and low in fibre a condition possibly found in Piperaceae [111,113]. Of all the treatments used, the bat species only exhibited a positive response to stimuli containing odor of ripe fruit. The experiments in this study revealed that C. perspicillata responded more frequently to P. hispidum while A. lituratus responded more frequently to Ficus insipida. Foraging activity of both species indicates a preference for particular fruit genera. Additionally, these bats showed favourable responses to olfactory stimuli, supporting the significance of olfaction in their foraging behaviour. In this regard, it appears that these bats may exclusively use olfactory cues to select and pick ripe food [93].
Previous studies proposed that, depending on the sensory or ecological niche to which a species has adapted, the significance of olfaction in bats changes. Studies examined whether the effects of ecological niche specialisation broadly observed among mammals [7] are associated with OR diversity over a more recent time frame in order to "fine-scale" our understanding of the relationship between OR evolution and ecological niche specialisation. This was completed by concentrating on the diversity in OR gene repertoire of the chiropteran radiation. In order to determine whether the evolution of the OR gene repertoire in bats was linked to sensory and ecological specialisation and to determine which gene families are significant in each ecological niche, OR gene repertoire was generated and compared across 27 bat species spanning the entire chiropteran phylogeny [3]. Researchers demonstrated that OR gene families OR 1/3/7 and OR 2/13 are related to the OR gene repertoire of frugivorous bats in two significant radiations: the Yangochiroptera, which includes New World fruit bats in the family Phyllostomidae, and the Yinpterochiroptera, which includes Old World fruit bats in the family Pteropodidae. Frugivorous bats have developed a liking for the fruit of A. muricata, whether it was native to the Old World or not [114]. It was speculated that the OR gene families OR 1/3/7 and OR 2/13 may be directly implicated in the detection of ethyl acetate given the relationship between these OR gene families and frugivorous Phyllostomids; however, this will need to be confirmed and investigated with subsequent functional experiments [3].
Researchers assumed that ecological specialization is expected to be connected to trait diversity, with generalist species exhibiting traits that enable access to a wider range of resources. This hypothesis states that these three bat species had overlapping geographic ranges, but with differing degrees of dietary specialization on Piper fruits [115]. To test this hypothesis, olfactory receptor genes of three closely related neotropical short-tailed fruit bats (Carollia castanea, C. sowelli and C. perspicillata) were sequenced using targeted sequence capture of probes designed from transcriptomic data to test whether specialist and generalist species had distinct receptor profiles. For each Carollia species, the number of intact OR genes were calculated and each subfamily of intact receptors were aligned (Bininda-Emonds, 2005). It was observed that OR gene families OR1/3/7 and OR 5/8/9 had twice the abundance when compared to the other OR gene families for all the species, while OR gene families OR55, OR12 and OR14 were represented by fewer paralogs comparative to the other gene families. C. perspicillata precisely had more ORs in subfamily OR5/8/9, but measures of diversity are quite similar across the three species (Lopez and Vaughan, 2007; Suzuki et al. 2018; Maynard et al 2019). Subfamily OR1/3/7 shows considerable differences in diversity among the three species even though C. sowelli and C. perspicillata have quite similar receptor counts [115].
Recent studies were conducted to measure the surface area of the olfactory epithelium distributed in the nasal cavity to check whether plant-visiting bats had abundant olfactory epithelia comparative to animal-feeding bats. μCT-scans of iodine-stained specimens were collected from 30 species with divergent diets [116]. There are typically five turbinate bones in the nasal cavity of Phyllostomidae and the majority of other Yangochiroptera species that have been previously investigated. These turbinate bones house the primary olfactory epithelium [91,117,118]. Interturbinal I was absent in Myotis albescens and Molossus rufus, but a small additional turbinal carrying olfactory epithelium was present towards the back of the olfactory recess. The congeneric Molossus molossus lacked this additional turbinal. Exploratory systems are frequently linked to evolvable genes and phenotypes because variation in these systems does not have the same potential fitness cost as it does in the core activities. Purifying selection replaces previously neutral processes when novel variable mutants are chosen in a specific niche. Environmental factors may then control that variety resulting from mutation [119]. We predicted a substantial correlation between molecular rates and morphological differences and plant visiting because we imagined a single expansion or shift to facilitate plant visiting (clear differences in plant-visiting bats independent of body size; would have revealed differences in rates of molecular evolution between plant feeders and animal feeders). Despite consistently high rates of molecular and morphological change, we discovered shorter OR molecular branch lengths in bats with higher epithelial surface areas [116].
The olfactory discrimination ability of Cynopterus sphinx to a variety of food odor substances were examined. Seven undiluted volatile odorant substances that are naturally present at various quantities in the food such as isoamyl acetate, ethyl acetate, hexanol, benzaldehyde, limonene, pinene, and dimethyl disulphide were used in the experiment for odor discrimination. Fruit pieces were offered as reward to the bats in addition to the odorant substances and the behavious of the bats were continuously recorded in an event recorder [120]. Except hexanol and dimethyl disulphide, more number of visits were made to the odorants when compared to the control. Among the odours, a gradational pattern was observed showing relatively maximum preference factor to ethyl acetate. Results suggest that C. sphinx is able to discriminate different food odor in a complex olfactory environment. The relatively higher number of visits and its subsequent decline to the control samples further emphasizes that the odors from our experimental samples play an important role on bat visits [120]. It was reported that C. sphinx detected fruits at short distances mainly by using olfactory cues, and our results suggest that in addition to detection of fruits C. sphinx could discriminate different odors at a short distance [3]. The results of current study validate an earlier report on Pteropus poliocephalus that displays olfactory discrimination between fruit derived and control odors at a decision distance of 125 mm [121].
In our study, the preliminary bioinformatics, molecular biology and sequencing results suggest that both fruit and insect eating bat species expressed different OR genes. Over all a total of 37 OR genes (9 gene families) were identified from 10 different bat species. We generated a global multiple alignment of the deduced amino acid sequences of 37 OR genes from 10 bat species. The most variable region was found to be TM segments 3, 4, and 5, within which 17 hypervariable regions were identified. These regions which constitute the odorant complementarity determining regions are the potential sites for ligand binding. Taken together, our preliminary results suggest that the total number of OR genes and families vary widely among both fruit and insect eating bats. The OR gene repertoire of bat species whose genomes have not yet been sequenced should be better sampled using next-generation sequencing techniques. These techniques would enable analysis of all OR genes amplified by defective primers. Additional research on bat OR genes would clarify their role and relationship to OR gene families. Also, the identification and cloning of functional OR repertoire lays the groundwork for attempting a number of outstanding problems in bat olfaction. Most importantly, it will ultimately contribute to understanding of structure-function correlations and small molecule recognition by this wide array of GPCRs in conjunction with reliable heterologous expression and assay techniques and high throughput screening of odorant libraries. Another fascinating subject is how genetic OR variation affects how different bat populations perceive odours differently. The evolution of the bat olfactory apparatus and its biological implications will be clarified by a comprehensive comparative examination of the functioning bat OR candidate gene and pseudogene repertoires. Our study represents a further step in revealing the function of bat OR genes and their associations to the gene families.
Conclusion
Recent years have seen a significant increase in our understanding of sensory systems as a result of laborious research into the mechanics underpinning olfaction. The revelation of the full human, mouse, and partial bat genome sequences, along wit
References
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Ann Rev Physiol. 2009:307-32.
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65(1):175-87.
- Hayden S, Bekaert M, Goodbla A, et al. A cluster of olfactory receptor genes linked to frugivory in bats. Mol Biol Evol. 2014;31(4):917-27.
- Niimura Y, Nei M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PloS One. 2007;2(8):e708.
- Keller A, Vosshall LB. Better smelling through genetics: mammalian odor perception. Curr Opin Neurobiol. 2008;18(4):364-9.
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9(12):951-63.
- Hayden S, Bekaert M, Crider TA, et al. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 2010;20(1):1-9.
- Niimura Y. Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr Genomics. 2012;13(2):103-14.
- Kang N, Koo J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012;45(11):612.
- Hildebrand JG, Shepherd GM. Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Ann Rev Neurosci. 1997;20:595.
- Eisthen HL. Why are olfactory systems of different animals so similar? Brain Behav Evol. 2002;59(5-6):273-93.
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5(4):263-78.
- Kosaka T, Kosaka K. Olfactory bulb anatomy.
- Sharma A, Kumar R, Ranjta S, et al. SMILES to smell: decoding the structure–odor relationship of chemical compounds using the deep neural network approach. J Chem Inf Model. 2021;61(2):676-88.
- Lohse MJ, Benovic JL, Codina J, et al. β-Arrestin: a protein that regulates β-adrenergic receptor function. Science. 1990;248(4962):1547-50.
- Rossiter KJ. Structure− odor relationships. Chem Rev. 1996;96(8):3201-40.
- Kermen F, Chakirian A, Sezille C, et al. Molecular complexity determines the number of olfactory notes and the pleasantness of smells. Sci Rep. 2011;1:206.
- Fleischer J, Breer H, Strotmann J. Mammalian olfactory receptors. Front Cell Neurosci. 2009:9.
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5(2):124-33.
- Abaffy T, Malhotra A, Luetje CW. The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues. J Biol Chem. 2007;282(2):1216-24.
- Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res. 2014;24(9):1485-96.
- Sosinsky A, Glusman G, Lancet D. The genomic structure of human olfactory receptor genes. Genomics. 2000;70(1):49-61.
- Young JM, Shykind BM, Lane RP, et al. Odorant receptor expressed sequence tags demonstrate olfactory expression of over 400 genes, extensive alternate splicing and unequal expression levels. Genome Biol. 2003;4(11):1-5.
- Ben-Arie N, Lancet D, Taylor C, et al. Olfactory receptor gene cluster on human chromosome 17: possible duplication of an ancestral receptor repertoire. Hum Mol Genet. 1994;3(2):229-35.
- Freitag J, Ludwig G, Andreini I, et al. Olfactory receptors in aquatic and terrestrial vertebrates. J Comp Physiol A. 1998;183(5):635-50.
- Glusman G, Yanai I, Rubin I, et al. The complete human olfactory subgenome. Genome Res. 2001;11(5):685-702.
- Abaffy T. Human olfactory receptors expression and their role in non-olfactory tissues-a mini-review. J Pharmacogenomics Pharmacoproteomics. 2015;6(4):1.
- Hughes GM, Boston ES, Finarelli JA, et al. The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol Biol Evol. 2018;35(6):1390-406.
- Tsuboi A, Miyazaki T, Imai T, et al. Olfactory sensory neurons expressing class I odorant receptors converge their axons on an antero dorsal domain of the olfactory bulb in the mouse. Eur J Neurosci. 2006;23(6):1436-44.
- Gaillard I, Rouquier S, Giorgi D. Olfactory receptors. Cell Mol Life Sci. 2004;61(4):456-69.
- Young JM, Trask BJ. V2R gene families degenerated in primates, dog and cow, but expanded in opossum. Trends Genet. 2007;23(5):212-5.
- Zhang X, Zhang X, Firestein S. Comparative genomics of odorant and pheromone receptor genes in rodents. Genomics. 2007;89(4):441-50.
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. PNAS. 2005;102(17):6039-44.
- Jacoby E, Bouhelal R, Gerspacher M, et al. The 7 TM G‐protein‐coupled receptor target family. ChemMedChem: Chemistry Enabling Drug Discovery. 2006;1(8):760-82.
- Liu AH, Zhang X, Stolovitzky GA, et al. Motif-based construction of a functional map for mammalian olfactory receptors. Genomics. 2003;81(5):443-56.
- Bhandawat V, Reisert J, Yau KW. Elementary response of olfactory receptor neurons to odorants. Science. 2005;308(5730):1931-4.
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48(3):417-30.
- Zufall F, Firestein S, Shepherd GM. Cyclic nucleotide-gated ion channels and sensory transduction in olfactory receptor neurons. Annu Rev Biophys. 1994;23(1):577-607.
- Reisert J, Bauer PJ, Yau KW, et al. The Ca-activated Cl channel and its control in rat olfactory receptor neurons. J Gen Physiol. 2003;122(3):349-64.
- Boekhoff I, Raming K, Breer H. Pheromone-induced stimulation of inositol-trisphosphate formation in insect antennae is mediated by G-proteins. J Comp Physiol B. 1990;160(1):99-103.
- Komatsu H, Jin YH, L'Etoile N, et al. Functional reconstitution of a heteromeric cyclic nucleotide-gated channel of Caenorhabditis elegans in cultured cells. Brain Res. 1999;821(1):160-8.
- Bobkov YV, Ache BW. Pharmacological properties and functional role of a TRP-related ion channel in lobster olfactory receptor neurons. J Neurophysiol. 2005;93(3):1372-80.
- Munger SD, Gleeson RA, Aldrich HC, et al. Characterization of a phosphoinositide-mediated odor transduction pathway reveals plasma membrane localization of an inositol 1, 4, 5-trisphosphate receptor in lobster olfactory receptor neurons. J Biol Chem. 2000;275(27):20450-7.
- Bruch RC, Teeter JH. Second-messenger signalling mechanisms in olfaction. InChemical Senses 2021 May 30 (pp. 283-298). CRC Press..
- Spehr M, Wetzel CH, Hatt H, et al. 3-phosphoinositides modulate cyclic nucleotide signaling in olfactory receptor neurons. Neuron. 2002;33(5):731-9.
- Cometto-Muñiz JE, Cain WS, Abraham MH. Quantification of chemical vapors in chemosensory research. Chem Senses. 2003;28(6):467-77.
- Su CY, Martelli C, Emonet T, et al. Temporal coding of odor mixtures in an olfactory receptor neuron. PNAS. 2011;108(12):5075-80.
- Martelli C, Carlson JR, Emonet T. Intensity invariant dynamics and odor-specific latencies in olfactory receptor neuron response. J Neurosci. 2013;33(15):6285-97.
- Szyszka P, Stierle JS. Mixture processing and odor-object segregation in insects. Prog Brain Res. 2014;208:63-85.
- Murlis J, Willis MA, Cardé RT. Spatial and temporal structures of pheromone plumes in fields and forests. Physiol entomol. 2000;25(3):211-22.
- Celani A, Villermaux E, Vergassola M. Odor landscapes in turbulent environments. Phys Rev X. 2014;4(4):041015.
- Vassar R, Ngai J, Axel R. Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell. 1993;74(2):309-18.
- Touhara K, Sengoku S, Inaki K, et al. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. PNAS. 1999;96(7):4040-5.
- Buck LB. Information coding in the mammalian olfactory system. Cold Spring Harb Symp Quant Biol. 1996;61:147-155).
- Vanderhaeghen P, Schurmans S, Vassart G, et al. Olfactory receptors are displayed on dog mature sperm cells. J Cell Biol. 1993;123(6):1441-52.
- Vanderhaeghen P, Schurmans S, Vassart G, et al. Molecular cloning and chromosomal mapping of olfactory receptor genes expressed in the male germ line: evidence for their wide distribution in the human genome. BBRC. 1997;237(2):283-7.
- Zhao H, Ivic L, Otaki JM, et al. Functional expression of a mammalian odorant receptor. Science. 1998;279(5348):237-42.
- Breer H. Olfactory receptors: molecular basis for recognition and discrimination of odors. Anal Bioanal Chem. 2003;377(3): 427–33.
- Wetzel CH, Oles M, Wellerdieck C, et al. Journal of Neuroscience. 1999; 19: 7426–7433.
- Kajiya K, Inaki K, Tanaka M, et al. Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21(16):6018-25.
- Malnic B, Hirono J, Sato T, et al. Combinatorial receptor codes for odors. Cell. 1999;96(5):713-23..
- Krautwurst D, Yau KW, Reed R. Cell. 1998; 95: 917–926.
- Shirasu M, Yoshikawa K, Takai Y, et al. Olfactory receptor and neural pathway responsible for highly selective sensing of musk odors. Neuron. 2014;81(1):165-78.
- Saito H, Chi Q, Zhuang H, et al. Odor coding by a Mammalian receptor repertoire. Sci Signal. 2009;2(60):ra9.
- Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413(6852):211-8.
- Matsui A, Go Y, Niimura Y. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol Biol Evol. 2010;27(5):1192-200.
- Cotton JA. The impact of gene duplication on human genome evolution. eLS. 2008.
- Chang D, Duda Jr TF. Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol Biol Evol. 2012;29(8):2019-29.
- Rizvanovic A, Amundin M, Laska M. Olfactory discrimination ability of Asian elephants (Elephas maximus) for structurally related odorants. Chem Senses. 2013;38(2):107-18.
- Touhara K, Vosshall LB. Sensing odorants and pheromones with chemosensory receptors. Ann Rev Physiol. 2009:307-32.
- Hughes GM, Gang L, Murphy WJ, et al. Using I llumina next generation sequencing technologies to sequence multigene families in de novo species. Mol Ecol Resour. 2013;13(3):510-21.
- Tautz D, Ellegren H, Weigel D. Next generation molecular ecology. Mol Ecol. 2010;19:1-3.
- Angeloni F, Wagemaker N, Vergeer P, et al. Genomic toolboxes for conservation biologists. Evol Appl. 2011;5:130–143.
- Alkan C, Sajjadian S, Eichler EE. Limitations of next-generation genome sequence assembly. Nat methods. 2011;8(1):61-5.
- Altringham JD, Fenton MB. Sensory ecology and communication in the Chiroptera. Bat ecology. 2003:90-127.
- Bates PJJ, Harrison DL. Bats of the Indian Subcontinent, Harrison Zoological Museum Press, England. 1997.
- Simmons NB. Bats Magazine. Bat Conservation International. Bats. 2015; 34: 1-23.
- Marshall AG. Bats, flowers and fruit: evolutionary relationships in the Old World. Biol J Linn Soc. 1983;20(1):115-35.
- Teeling EC, Springer MS, Madsen O, et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307(5709):580-4.
- Zhao H, Rossiter SJ, Teeling EC, et al. The evolution of color vision in nocturnal mammals. PNAS. 2009;106(22):8980-5.
- Zhang G, Li C, Li Q, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346(6215):1311-20.
- Zhang Q, Edwards SV. The evolution of intron size in amniotes: a role for powered flight? Genome Biol Evol. 2012;4(10):1033-43.
- Zhang G, Cowled C, Shi Z, et al. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 2013;339(6118):456-60.
- Tsagkogeorga G, Müller S, Dessimoz C, et al. Comparative genomics reveals contraction in olfactory receptor genes in bats. Sci Rep. 2017;7(1):1-0.
- Thies W, Kalko EK, Schnitzler HU. The roles of echolocation and olfaction in two Neotropical fruit-eating bats, Carollia perspicillata and C. castanea, feeding on Piper. Behav Ecol Sociobiol. 1998;42(6):397-409.
- Korine C, Kalko EK. Fruit detection and discrimination by small fruit-eating bats (Phyllostomidae): echolocation call design and olfaction. Behav Ecol Sociobiol. 2005;59(1):12-23.
- Laska M. Olfactory discrimination ability in short-tailed fruit bat, Carollia perspicillata (Chiroptera: Phyllostomatidae). J Chem Ecol. 1990;16(12):3291-9.
- Brokaw AF, Davis E, Page RA, et al. Flying bats use serial sampling to locate odour sources. Biol Lett. 2021;17(10):20210430.
- Handley Jr CO, Wilson DE, Gardner AL. Demography and natural history of the common fruit bat, Artibeus jamaicensis, on Barro Colorado Island, Panama.
- Ortega J, Castro-Arellano I. Artibeus jamaicensis. Mammalian species. 2001;2001(662):1-9.
- Bhatnagar KP, Kallen FC. Morphology of the nasal cavities and associated structures in Artibeus jamaicensis and Myotis lucifugus. Am J Anat. 1974;139(2):167-89.
- Hodgkison R, Ayasse M, Häberlein C, et al. Fruit bats and bat fruits: the evolution of fruit scent in relation to the foraging behaviour of bats in the New and Old World tropics. Funct Ecol. 2013;27(4):1075-84.
- Parolin LC, Mikich SB, Bianconi GV. Olfaction in the fruit-eating bats Artibeus lituratus and Carollia perspicillata: an experimental analysis. An Acad Bras Cienc. 2015;87:2047-53.
- da Costa RS, Bicca-Marques JC. Owl monkeys (Aotus nigriceps and A. infulatus) follow routes instead of food-related cues during foraging in captivity. PloS One. 2014;9(12):e115188.
- Gire DH, Kapoor V, Arrighi-Allisan A, Seminara A, Murthy VN. Mice develop efficient strategies for foraging and navigation using complex natural stimuli. Curr Biol 2016;26(10):1261-73.
- Bohn KM, Moss CF, Wilkinson GS. Pup guarding by greater spear-nosed bats. Behav Ecol Sociobiol. 2009;63(12):1693-703.
- Watkins LC, Shump Jr KA. Behavior of the evening bat Nycticeius humeralis at a nursery roost. Am Midl Nat 1981:258-68.
- Gustin MK, McCracken GF. Scent recognition between females and pups in the bat Tadarida brasiliensis mexicana. Anim Behav. 1987;35(1):13-9.
- Wierucka K, Pitcher BJ, Harcourt R, et al. Multimodal mother–offspring recognition: the relative importance of sensory cues in a colonial mammal. Anim Behav. 2018;146:135-42.
- Bloss J. Olfaction and the use of chemical signals in bats. Acta Chiropterologica. 1999;1(1).
- Liang J, Yang J, Chen Y, et al. The role of olfactory cues in mother–pup, groupmate, and sex recognition of lesser flat‐headed bats, Tylonycteris pachypus. Ecol Evol. 2021;11(22):15792-9.
- Brokaw AF, Smotherman M. Role of ecology in shaping external nasal morphology in bats and implications for olfactory tracking. PloS One. 2020;15(1):e0226689.
- Von Helversen O, Winkler L, Bestmann HJ. Sulphur-containing “perfumes” attract flower-visiting bats. J Comp Physiol A. 2000;186(2):143-53.
- Mikich SB, Bianconi GV, Maia BH, et al. Attraction of the fruit-eating bat Carollia perspicillata to Piper gaudichaudianum essential oil. J Chem Ecol. 2003;29(10):2379-83.
- Carter GG, Ratcliffe JM, Galef BG. Flower bats (Glossophaga soricina) and fruit bats (Carollia perspicillata) rely on spatial cues over shapes and scents when relocating food. PloS One. 2010 May 25;5(5):e10808.
- Morrison DW. Influence of habitat on the foraging distances of the fruit bat, Artibeus jamaicensis. J Mammal. 1978;59(3):622-4.
- Mikich SB. A dieta dos morcegos frugívoros (Mammalia, Chiroptera, Phyllostomidae) de um pequeno remanescente de Floresta Estacionai Semidecidual do sul do Brasil. Rev Bras Zool. 2002;19:239-49.
- Olea-Wagner A, Lorenzo C, Naranjo E, et al. Diversity of fruits consumed by three species of bats (Chiroptera: Phyllostomidae) in the Lacandona rainforest, Chiapas, Mexico. Rev Mex Biodivers. 2007;78(1):191-200.
- Bonaccorso FJ, Gush TJ. Feeding behaviour and foraging strategies of captive phyllostomid fruit bats: an experimental study. J Anim Ecol. 1987:907-20.
- Dumont ER. An Ecomorphological Approach. Bat ecology. 2003:398.
- Fleming TH. The short-tailed fruit bat: a study in plant-animal interactions. University of Chicago press; 1988.
- Charles-Dominique P. Feeding strategy and activity budget of the frugivorous bat Carollia perspicillata (Chiroptera: Phyllostomidae) in French Guiana. J Trop Ecol. 1991;7(2):243-56.
- Thies W, Kalko EK. Phenology of neotropical pepper plants (Piperaceae) and their association with their main dispersers, two short‐tailed fruit bats, Carollia perspicillata and C. castanea (Phyllostomidae). Oikos. 2004;104(2):362-76..
- Spencer HJ, Fleming TH. Roosting and Foraging Behavior of the Queensland Tube-Nosed Bat, Nyctimene-Robinsoni (Pteropodidae)-Preliminary Radio-Tracking Observations. Wildl Res. 1989;16(4):413-20.
- Yohe LR, Leiser-Miller LB, Kaliszewska ZA, et al. Diversity in olfactory receptor repertoires is associated with dietary specialization in a genus of frugivorous bat. G3. 2021;11(10):jkab260.
- Yohe LR, Fabbri M, Lee D, et al. Ecological constraints on highly evolvable olfactory receptor genes and morphology in neotropical bats. Evol. 2022;76(10):2347-60.
- Bhatnagar KP, Kallen FC. Morphology of the nasal cavities and associated structures in Artibeus jamaicensis and Myotis lucifugus. Am J Anat. 1974;139(2):167-89.
- Yohe LR, Hoffmann S, Curtis A. Vomeronasal and olfactory structures in bats revealed by DiceCT clarify genetic evidence of function. Front Neuroanat. 2018:32.
- Kirschner M, Gerhart J. Evolvability. PNAS. 1998;95(15):8420-7.
- Elangovan V, Satya Priya EY, Marimuthu G. Olfactory discrimination ability of the short-nosed fruit bat Cynopterus sphinx. Acta Chiropt. 2006;8(1):247-53.
- Oldfield AC, Tidemann CR, Robinson AP. Olfactory discrimination in the Australian flying-foxes, Pteropus poliocephalus and P. scapulatus (Chiroptera: Pteropodidae). Bat Research News. 1993;34:33.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref