Research Article - Microbiology: Current Research (2018) Volume 2, Issue 2
Molecular identification of a Begomovirus associated with yellow vein net disease on Malva parviflora L. from india.
Snehi SK1*, Parihar SS1, Gupta G1, Purvia AS2 and Singh V1
1Department of Microbiology, Barkatullah University, Bhopal-462026, M.P., India
2Department of Biology, Virangna Uda Devi Government Girls Inter College, Mall, Lucknow-226104, U.P., India
- *Corresponding Author:
- Sunil kumar Snehi
Department of Microbiology
Barkatullah University
Bhopal-462026,M.P., India
E-mail: sunilsnehi@gmail.com
Accepted date: January 31, 2018
Citation: Snehi SK, Parihar SS, Gupta G, et al. Molecular identification of a Begomovirus associated with yellow vein net disease on Malva parviflora L. from india. Microbiol Curr Res. 2018;2(2):24-29.
DOI: 10.4066/2591-8036.17-3895
Visit for more related articles at Microbiology: Current ResearchAbstract
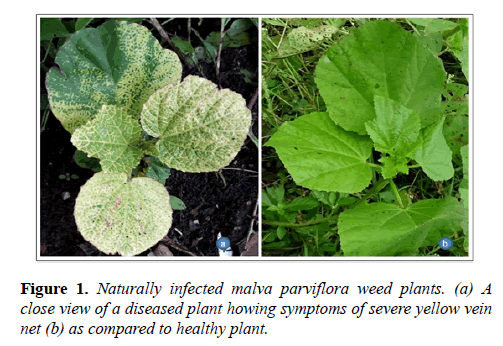
Incidence of yellow vein net disease with leaf distortion was observed on Malva parviflora grown as a weed in Barkatullah University campus, Bhopal, India during the rainy session. The begomovirus disease was suspected on the basis of symptomatology and whiteflies insects’ population on the plant. The begomovirus was detected by the PCR with the begomovirus gene specific primers. The begomovirus under study showed highest nucleotide sequence identities and distinct phylogenetic relationships of coat protein gene (CP) with several isolates of Tomato leaf curl Kerala virus (ToLCKeV). This is the first report of ToLCKeV associated with yellow vein net disease on M. parviflora and it is a new host of begomovirus from India.
Keywords
Malva parviflora, begomovirus, Yellow vein net disease, Sequence identities, and Tomato leaf curl Kerala virus
Introduction
Malva parviflora L. (Family Malvaceae) commonly known as cheese weed and it is an exotic annual weed or perennial herb that is native to Northern Africa, Europe and Asia. It is used in the treatment of cough, throat infection and other bronchial problems as well as stomach and intestine irritations. The flowers and leaves are emollient and used for the softening of sensitive area of the skin. Combine with Eucalyptus; it makes a good remedy for cough and other chest ailments [1].
Madhya Pradesh is a central region of India and agriculture is one of the main sectors of the state’s economy. About 73 percent population of the state is rural, which directly or indirectly depends on the agriculture. The Madhya Pradesh has plant diversity and flexible temperatures, which are favourable to the virus insect vector (aphids, whiteflies, leafhoppers, and planthoppers). There are few reports has been reported of the begomoviruses infection on Jatropha gossypifolia [2], Solanum lycopersicum [3] and Cnidoscolus acontifolia [4] from Madhya Pradesh state, India.
Begomoviruses is the largest genera in the family Geminiviridae. Begomoviruses have a circular, single-stranded deoxyribonucleic acid (ssDNA) genome and are transmitted in nature by the whitefly (Bemisia tabaci), and causes significant yield losses in economically important crop plants worldwide [5,6]. Begomoviruses generally have bipartite genomes (designated as DNA-A and DNA-B) and infect dicotyledonous plants. Based on their genome characteristics and phylogenetic relationships, Begomoviruses have been divided broadly into Old World (OW) viruses (Eastern hemisphere, Europe, Africa, Asia and Australasia) and the New World (NW) viruses (Western hemisphere, the Americas) [7,8]. Monopartite begomoviruses (have DNA-A genome only) are predominantly found in the Old World and are often associated with satellite DNAs (alpha-and betasatellites), which may or may not contribute to pathogenicity [9].
The first reported of the Malva veinal necrosis virus considered as belonging to the potato X virus group in Malva parviflora from Brazil [10]. In Israel, natural infection of Tomato yellow leaf curl virus-Isreal (TYLCV-Is) was found in the annual weed M. Paraviflora [11]. M. parvifolia acts as host for many viruses including the South African Cassava mosaic virus [12] and Faba bean necrotic yellow virus in Jordan [13]. In 2003, Antignus et al. reported that Squash leaf curl virus (SLCV) could infect Malva nicaeensis and Ecballium elaterium (Cucurbitaceae) in Israel [14]. Squash leaf curl virus (SLCV) was also found to occur naturally in M. parviflora, with severe leaf curling, yellowing and stunting of the whole plants. The full-length genomes of Squash leaf curl virus-Malva (SLCV-Malva) isolate were amplified using the bacteriophage F DNA polymerase enzyme [15]. Recently, the identified full-length begomovirus genome shared maximum nucleotide (nt) sequence identity at 92.5% with Hollyhock leaf curl virus (HoLCV), representing a new strain, Ageratum conyzoides symptomless alphasatellite (ACSLA) and Ageratum yellow vein India alphasatellite (AYVIA), new isolates of ACSLA and AYVIA identified from M. parviflora in Pakistan [16].
We report here, molecular detection and identification of a Tomato leaf curl Kerala virus associated with yellow vein net disease on M. parviflora from central region of Madhya Pradesh of India, based on sequence analysis of complete coat protein gene.
Materials and Methods
Virus source
Naturally infected M. parviflora plants showing severe yellow vein net disease with leaf distortion were collected from Barkatullah University campus, Bhopal, during rainy session (July) in year of 2016.
DNA extraction and polymerase chain reaction (PCR)
The total DNA was extracted from symptomatic and asymptomatic leaf samples of M. parviflora plants by Dellaporta et al. [17] method. To detection of begomovirus, the polymerase chain reactions (PCR) were carried out using total DNA as template and a set of begomovirus coat protein gene specific primers CPIT-I/CPIT-T (Acc. AM180920 and AM180921). The PCRs were set up in a 50 µL reaction mixture containing: template DNA (100 mg), dNTPs (10 mM each), primers (each 25 pmol), Taq DNA polymerase (1.5 U, Merk Pvt. Ltd), assay buffer 10X (Merk Pvt. Ltd) in a thermal cycler (Bio-Rad, USA). The PCR was done with the conditions: initial denaturation at 94°C for 5 min, followed by followed by 30 cycles of denaturation at 94°C for 1 min; annealing temperatures for 1 min at 47°C, extension at 72°C for 1.5 min and final extension cycle was 5 min at 72°C.
Sequence analysis of viral gene
The sequence data from the virus isolate under study were analysed by BLASTn (http:// www.ncbi.nlm.nih.gov/BLAST) and compared with existing sequences of begomovirus species available in the GenBank database. ORFs were translated into amino acids using the Expasy tool (http://www.expasy. org/tools/dna.html). The sequence similarities of selected begomovirus species were obtained using the Genomatix DiAlign 2 program based on pair-wise alignment. Phylogenetic analyses were performed using the Molecular Evolutionary Genetics Analysis tool (MEGA v.7.1,) with 1000 replicates bootstrapping and the tree was generated with the Neighbour joining method.
Results
Disease symptoms
During a survey in rainy session July 2016, the begomovirus-like symptoms were observed on a large number of M. parviflora plants growing in Barkatullah University campus, Bhopal Madhya Pradesh with the disease incidence about 40%. The naturally infected plants exhibited severe yellow mosaic and leaf distortion symptoms (Figure 1). A population of whiteflies (Bemisia tabaci) was also noticed in the growing area therefore, association of begomovirus with the disease was suspected.
Amplification of the begomovirus by PCR
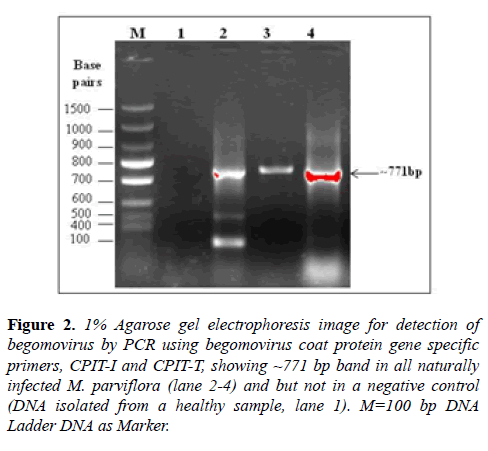
The begomovirus was detected by PCR reactions using the total DNA as template and begomovirus coat protein gene specific primers: CPIT-I/CPIT-T. During PCR, the expected bands of ~800 bp were amplified from (3/3) naturally infected symptomatic plant samples but not in a healthy sample 1 (Figure 2). The PCR products (three samples) were purified by using PCR Clean-up System Kit (Promega, USA) and purified PCR product was sequenced. The consensus sequence data of three identical sequences were analyzed by the BLASTn and complete coat protein gene of 771 nucleotides of begomovirus under study isolate was deposited to GenBank database (Accession KY511140).
Figure 2: 1% Agarose gel electrophoresis image for detection of begomovirus by PCR using begomovirus coat protein gene specific primers, CPIT-I and CPIT-T, showing ~771 bp band in all naturally infected M. parviflora (lane 2-4) and but not in a negative control (DNA isolated from a healthy sample, lane 1). M=100 bp DNA Ladder DNA as Marker.
Sequence analysis and Phylogeny of under study virus isolate
BLASTn analysis of the coat protein gene of M. parviflora (KY511140) of the begomovirus isolate from M. parviflora revealed highest 95-98% sequence identity with several isolates of Tomato leaf curl Kerala virus (ToLCKeV) on Solanum lycopersicum (KF551575, EU910140, EU910141, LT556075, LN886521, KY216063) and Brassica rapa (KX671963) from India and Pakistan. The isolate also shared 94% identities with isolates of Pedilanthus leaf curl virus (PeLCV: LT795118, LT795117) on Brassica rapa from Pakistan and 90-91% sequence identities with other begomovirus isolates.
During Genomatix DiAlign analysis of coat protein gene of (KY511140) with the respective sequences of selected begomoviruses, the virus isolate under study showed highest 93-97% similarities at nucleotide (nt) level and 95-97% similarity at amino acid (aa) level with Tomato leaf curl Kerala virus (ToLCKeV) on Solanum lycopersicum from India and Pakistan (GQ924760, JF496657). The isolate also shared 84-91% at nt level and 87-96% similarties with aa level with other selected begomoviruses: PeLCV, ChLCV, ChLCMV, ToLCKV, CheToLCV, ToLCV (Table 1).
| Accessions | Virus | Host | Country | %Identity Nucleotide | %Identity Amino Acid |
|---|---|---|---|---|---|
| KF551575 | ToLCKeV | Solanum lycopersicum | India | 97 | 98 |
| EU910140 | ToLCKeV | Solanum lycopersicum | India | 96 | 98 |
| EU910141 | ToLCKeV | Solanum lycopersicum | India | 95 | 96 |
| LT556075 | ToLCKeV | Solanum lycopersicum | Pakistan | 95 | 96 |
| KX671963 | ToLCKeV | Brassica rapa | Pakistan | 94 | 95 |
| LN886521 | ToLCKeV | Solanum lycopersicum | Pakistan | 93 | 96 |
| KY216063 | ToLCKeV | Solanum lycopersicum | India | 93 | 97 |
| LT795118 | PeLCV | Brassica rapa | Pakistan | 91 | 96 |
| LT795117 | PeLCV | Brassica rapa | Pakistan | 91 | 96 |
| KP195266 | ChLCMV | Solanum lycopersicum | India | 88 | 95 |
| KU760802 | ToLCV | Solanum melongena | India | 87 | 94 |
| KX246859 | ToLCKaV | Cajanus cajan | India | 87 | 91 |
| LN906594 | CheToLCV | Parthenium hysterophorus | Pakistan | 87 | 90 |
| LN906593 | CheToLCV | Parthenium hysterophorus | Pakistan | 86 | 87 |
| KX831454 | ToLCV | Ocimum basilicum | India | 86 | 91 |
| KP195261 | ToLCKaV | Solanum lycopersicum | India | 86 | 89 |
| AF336806 | ChLCV | Capsicum annuum | Pakistan | 84 | 94 |
| LN886660 | ChLCV | Capsicum annuum | Pakistan | 84 | 93 |
Table 1: Sequence identities of virus isolate of Malva parviflora (KY511140) with other selected begomovirus isoaltes at nucleotide (nt) and their at amino acid (aa) levels based on genomatix DiAlign programme. (Abbreviation of virus name: Tamato leaf Kerala virus; PeLCV: Pedilanthus leaf curl virus; ChLCV: Chilli leaf curl virus; chLCMV: Chilli leaf curl Multan virus; ToLCKV: Tamato leaf curl Karnataka virus; CheToLCV: Cherry Tamato leaf cuel virus; ToLCV: Tamato leaf cuel virus).
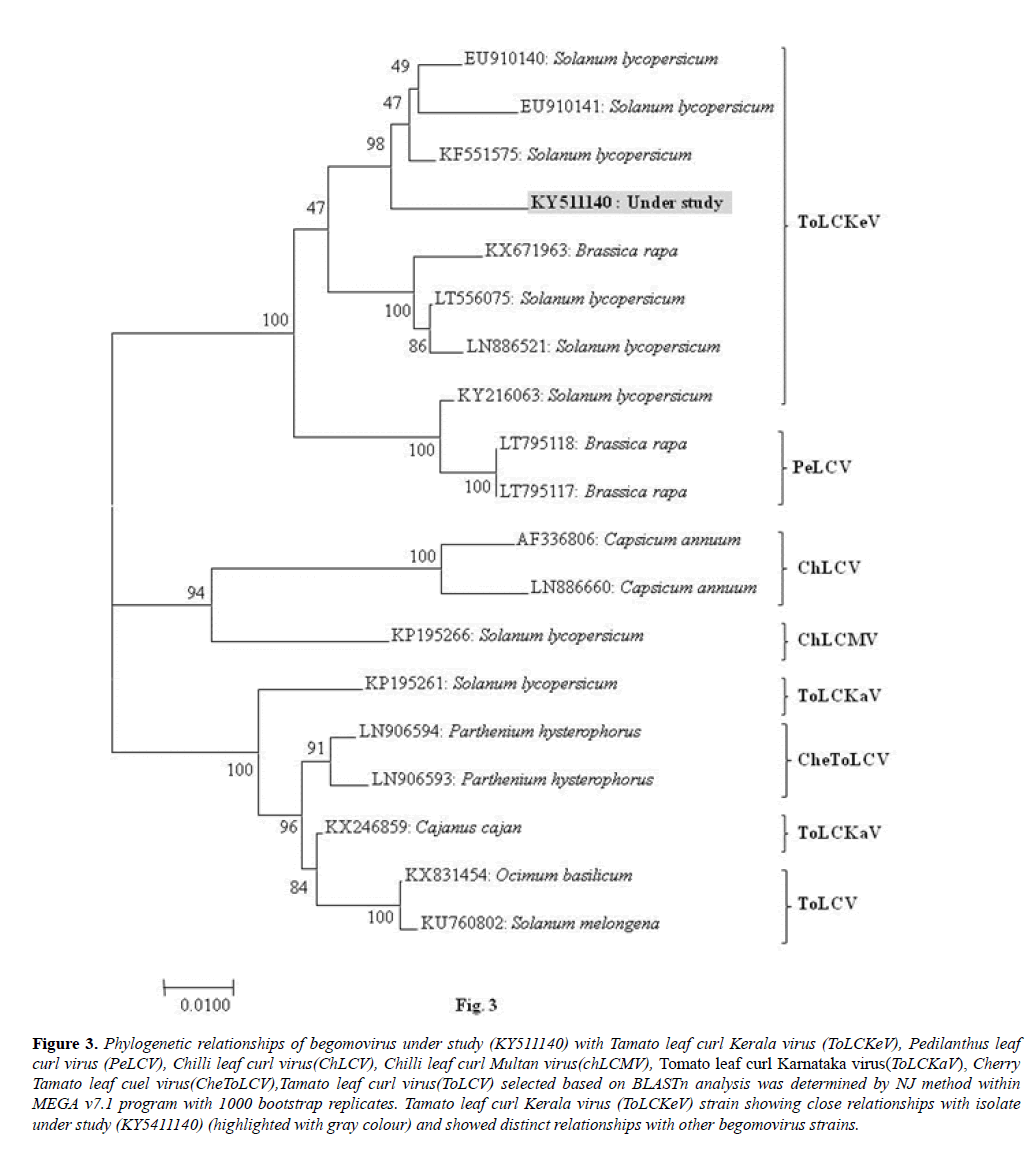
During phylogenetic analysis of virus isolate under study (KY511140) with the other begomovirus isolates was done using the Molecular Evolutionary Genetics Analysis Tool (MEGA v.7.1) with 1000 replicates of bootstrapping, and a dendrogram was generated by the neighbour-joining method and viewed using the NJ plot program.
The virus isolate (KY511140) shared close phylogenetic relationships with the isolate of Tomato leaf curl Kerala virus (ToLCKeV: KF551575, EU910140, EU910141, LT556075, LN886521, KY216063) and shared distinct relationships with the isolates of Pedilanthus leaf curl virus (PeLCV: LT795118 and LT795117); Chilli leaf curl virus (ChLCV: AF336806, LN886660) on chilli from Pakistan, Chilli leaf curl Multan virus (ChLCMV: KP195266) from tomato from India, Tomato leaf curl Karnataka virus (ToLCKaV: KP195261, KX246859) on Solanum lycopersicum and Cajanus cajan from India; Cherry tomato leaf curl virus (CheToLCV: LN906594, LN906593) on Parthenium hysterophorus from Pakistan and Tomato leaf curl virus (ToLCV: KX831454, KU760802) on Ocimum basilicum and Solanum melongena from India (Figure 3).
Figure 3: Phylogenetic relationships of begomovirus under study (KY511140) with Tamato leaf curl Kerala virus (ToLCKeV), Pedilanthus leaf curl virus (PeLCV), Chilli leaf curl virus(ChLCV), Chilli leaf curl Multan virus(chLCMV), Tomato leaf curl Karnataka virus(ToLCKaV), Cherry Tamato leaf cuel virus(CheToLCV),Tamato leaf curl virus(ToLCV) selected based on BLASTn analysis was determined by NJ method within MEGA v7.1 program with 1000 bootstrap replicates. Tamato leaf curl Kerala virus (ToLCKeV) strain showing close relationships with isolate under study (KY5411140) (highlighted with gray colour) and showed distinct relationships with other begomovirus strains.
On the basis of highest sequence identity and the close phylogenetic relationships of the virus isolate of M. parviflora with the corresponding sequences of various begomovirus isolates reported worldwide, the virus associated with yellow vein net disease of of M. parviflora was identified as a begomovirus isolate of Tomato leaf curl Kerala virus from Madhya Pradesh, India.
Discussion
India is a large country with very diverse agro-climatic conditions. On the one hand this diverse agro-climate makes India one of the richest sources of flora and fauna in the world, but on the other hand it creates ideal conditions for plant viruses. Although it has been established that weeds can play an important role in the emergence of plant viral epidemics affecting crops in different parts of the world [18,19], they are still neglected and only limited work has been carried out to characterize the begomovirus complexes associated with different weed species in India.
The occurrence begomovirus on weed plants have been reported from India like: Ageratum enation virus (AgEV) on Cleome gynandra [20], Crassocephalum crepidioides and Ageratum conyzoides [21]. A new begomovirus Rhynchosia yellow mosaic India virus associated with yellow mosaic disease in Rhynchosia minima [22]. Jatropha gossypifolia weed plants have been reported from the natural occurrence of Croton yellow vein mosaic virus and Croton yellow vein mosaic betasatellite from India [23] and a new begomovirus Jatropha yellow mosaic India virus also have been reported from J. gossypifolia [2].
Tomato Leaf Curl Kerala Virus (ToLCKeV), a virus prevalent in the tomato crop of Kerala state of India [24,25] and Alternanthera sessilis a new host of ToLCKeV have been reported from Rajasthan state, India [26].
In this study the association of the begomovirus with severe yellow vein net disease of M. parviflora (weed plant) was detected by PCR from using total DNA extracted from symptomatic leaf samples using coat protein gene specific primers for members of the genus Begomovirus, which revealed positive amplification of the expected-size bands (~800 bp). On the basis of positive PCR amplification, sequence analysis and phylogenetic relationships, the virus isolate was identified as a begomovirus that is closely related to the isolates of Tomatro leaf curl Kerala virus.
M. parvifolia acts as host for some viruses including Malva veinal necrosis virus is considered as belonging to the potato X virus group [10], Tomato yellow leaf curl virus- Isreal [11], South African Cassava mosaic virus [12], Faba bean necrotic yellow virus [13]). The Squash leaf curl virus from Israel was also found to occur naturally in M. parviflora [14] and Squash leaf curl virus-Malva from Jordan Valley [15]. Recently, Hollyhock leaf curl virus was identified on M. parviflora from Pakistan [16]. However, there are no reports have been published in literature from India about any kind of plant viruses including begomovirus on M. parvifolia plant.
We report here that the natural occurrence of a begomovirus associated with yellow vein net disease of M. parviflora identified as a new host of begomovirus isolate of Tomato leaf curl Kerala virus from Madhya Pradesh, India. Because M. parviflora grows as a weed in India and abroad near the agricultural fields, so that the associated Tomato leaf curl Kerala virus is a serious threat to other commercially important crops and may contribute to the epidemiology of Tomato leaf curl Kerala virus diseases in India.
Acknowledgement
Authors are thankful to the Vice Chancellor, Barkatullah University, Bhopal, M.P. for facilities and UGC, New Delhi for funding under UGC-BSR Research Start-Up Grant project.
References
- Benzie IF, Wachtel-Galor S. Herbal medicine: biomolecular and clinical aspects, oxidative stress and disease(2nd ed)Florida: CRC Press. 2011;499.
- Snehi SK, Raj SK, Khan MS, et al. Molecular identification of a new Begomovirus species associated with yellow mosaic disease of Jatropha gossypifolia in India. Arch Virol. 2011;156:2303–7.
- Snehi SK, Parihar SS, Gupta G, et al. Molecular Detection and Identification of Begomovirus Isolate on Tomato from Central Region of India. J Plant Pathol Micro. 2016;7:389.
- Snehi SK, Purvia AS, Gupta G, et al. Molecular detection of a Begomovirus species on Chaya (Cnidoscolus acontifolia) from Madhya Pradesh, India which is distantly related to Sri Lankan cassava mosaic virus. Virol Mycol. 2017;6:1.
- Varma A, Malathi VG. Emerging geminivirus problems: A serious threat to crop production. Anna Appl Biol. 2003;142:145-6.
- Stanley J, Bisaro DM, Briddon RW, et al. Virus Taxonomy, VIIIth Report of the ICTV. Elsevier/Academic Press, London. 2005;301-26.
- Rybicki EP. A phylogenetic and evolutionary justification for three genera geminiviridae. Arch Virol. 1994;139:49-78.
- Nawaj-ul Rehman MS, Fauquet CM. Evolution of geminiviruses and their satellites. FEBS Lett. 2009;583:1825-32.
- Briddon RW, Brown JK, Moriones E, et al. Recommendations for the classification and nomenclature of the DNA-β satellites of begomoviruses. Arch Virol. 2008;153:763–81.
- Costa AS, Kitajima EW. Veinal necrosis, a malva disease caused by a virus of the potato X group. Bragantia. 1970;29:51.
- Cohen S, Kern J, Harpaz I, et al. Epidemiological studies of the Tomato leaf curl virus (TYLCV) in the Jorden Valley, Isreal. Phytoparasitica. 1998;16:259-70.
- Berrie LC, rybicki EP, Rey MEC. Complete nucleotide sequence and host range of South Africa cassava mosaic virus: future evidence for recombination amongst begomoviruses. J Gen Virol. 2001;82:53-8.
- Al-Nsour A, Mansour A, Al-Musa A, et al. Distribution and incidence of Faba bean necrotic yellows virus in Jordan. Plant Pathol.1998;47:510–5.
- Antignus Y, Lachman O, Pearlsman M, et al. Squash leaf curl geminivirus- a new illegal immigrant from the Western Hemisphere and a threat to cucurbit crops in Israel. Phytoparasitica. 2003;31:415.
- Al-Musa A, Anfoka G, Misbeh S, et al. Detection and Molecular Characterization of Squash leaf curl virus (SLCV) in Jordan. J Phytopathol. 2008;156:311–6.
- Sattar MN, Qurashi F, Iqbal Z, et al. Molecular characterization of Hollyhock leaf curl virus and associated DNA-satellites infecting Malva parviflora in Pakistan. Can J Plant Pathol. 2017;39:229-34.
- Dellaporta SL, Wood JJ, Hicks JB. A plant DNA mini-preparation: version II. Plant Mol Biol Rep. 1983;1:19-21.
- McLaughlin PD, McLaughlin WA, Maxwell DP, et al. Identification of begomoviruses infecting crops and weeds in Belize. Plant Viruses. 2008;2:58-63.
- Rojas A, Kvarheden A, Valkonen JPT. Geminiviruses affecting tomato crops in Nicaragua. Plant Disease. 2000;84:843-6.
- Raj SK, Snehi SK, Khan MS, et al. Detection of Ageratum enation virus from Cat’s whiskers (Cleome gynandra L.) with leaf curl symptoms in India. J Gen Plant Pathol. 2010;76:292-4.
- Kumar Y, Hallan V, Zaidi AA. First report of Ageratum enation virus infecting Crassocephalum crepidioides (Benth.) S. Moore and Ageratum conyzoides L. in India. J Gen Plant Pathol. 2011;77:214-6.
- Jyothsna P, Rawat R, Malathi VG. Molecular characterization of a new begomovirus infecting a leguminous weed Rhynchosia minima in India. Virus Genes. 2011;42:407-14.
- Snehi SK, Khan MS, Raj SK, et al. Complete nucleotide sequence of Croton yellow vein mosaic virus and DNA-β associated with yellow vein mosaic disease of Jatropha gossypifolia in India.Virus Genes. 2011;43:93-101.
- Pasumarthy KK, Choudhury NR, Mukherjee SK. Tomato leaf curl Kerala virus (ToLCKeV) AC3 protein forms a higher order oligomer and enhances ATPase activity of replication initiator protein (Rep/AC1). J Virol. 2010;7:128.
- Pandey P, Mukhopadhya S, Naqvi AR, et al. Molecular characterization of two distinct monopartite begomoviruses infecting tomato in India. Virology J. 2010;7:337.
- Marwal A, Sahu AK, Prajapat R, et al. Molecular and Recombinational Characterization of Begomovirus Infecting an Ornamental Plant Alternanthera sessilis: A new host of Tomato Leaf Curl Kerala Virus reported in India. Science International. 2013;1:51-6.