Research Article - Biomedical Research (2018) Volume 29, Issue 6
MicroRNA-34b attenuates proliferation and invasion of osteosarcoma cells by directly targeting CCND1
Yu Hou1, Jianbao Jiao1, Bo Sun1, Taoping Chen1, Zhao Guo1, Yunfei Wang1, Helin Feng2 and Zhixing Liang1
1Department of Orthopaedics, the Affiliated Hospital of Hebei University, Hebei, PR China
2Department of Orthopaedics, the Fourth Hospital of Hebei Medical University, Hebei, PR China
- *Corresponding Author:
- Zhixing Liang
Department of Orthopaedics
The Affiliated Hospital of Hebei University
PR China
Accepted date: December 20, 2017
DOI: 10.4066/biomedicalresearch.29-17-3724
Visit for more related articles at Biomedical ResearchAbstract
MicroRNAs (miRNAs) deregulation has been reported to play important roles in tumorigenesis and tumor development through the regulation of their targets in Osteosarcoma (OS). Hence, fully understanding the biological roles and mechanisms of OS-related miRNAs may contribute to the development of novel therapeutic targets for patients with this disease. In this study, we found that miR-34b expression was downregulated in OS tissues and cell lines. The resumption of miR-34b expression led to a significant reduction in cell proliferation and invasion of OS. Meanwhile, Cyclin D1 (CCND1) was identified as a direct target of miR-34b in OS through bioinformatics analysis, luciferase reporter assay, quantitative reverse-transcription PCR (RT-qPCR) and Western blot analysis. The results of the present study demonstrated that low miR-34b expression is involved in OS formation and progression, suggesting that miR-34b/CCND1 axis may serve as a potential therapeutic target in OS oncogenesis.
Keywords
Osteosarcoma, microRNA-34b, Proliferation, Invasion, Cyclin D1
Introduction
Osteosarcoma (OS), a common malignant primary bone tumor, mainly affects the long bones of the legs and arms with a high fatality rate in children and adolescents [1]. In recent years, several risk factors have been identified to contribute to OS formation and progression. These factors include ionising radiation, alkylating agents, Paget’s disease, hereditary retinoblastoma and genetic aberrations [2]. Despite the current advances in the treatment approaches for OS, the clinical outcomes and prognosis remain unsatisfactory. Recurrence and metastasis are the major reasons for poor prognosis [3]. Therefore, elucidating the mechanisms underlying OS occurrence and development is urgently needed to investigate novel therapeutic methods for patients with such malignancy.
MicroRNAs (miRNAs) are a large group of short non-coding single-stranded RNA molecules that negatively regulate gene expression through partial complementary binding to the 3’- Untranslated Regions (UTRs) of their target genes. This interaction results in mRNA degradation and/or translation inhibition [4]. Bioinformatics analysis indicated that miRNAs, which account for approximately 1% of all human genes, can modulate approximately 60% of human protein-coding genes [5]. Substantial evidence has shown that miRNAs play key roles in various biological processes, including cell proliferation, apoptosis, invasion, metabolism and metastasis [6]. MiRNAs have been reported to be aberrantly expressed in almost all human cancer types. Deregulated miRNAs may serve as oncogenes or tumor suppressors in tumorigenesis and tumor development mainly depending on the biological characteristics of their target genes [7]. Therefore, elucidating the OS-related miRNAs may provide novel and effective therapeutic target to treat this disease.
MiR-34b is regularly abnormally expressed in numerous human cancer types. MiR-34b dysregulation has been reported in several human malignancy types. For example, miR-34b expression was silenced in breast cancer tissues and cell lines. Low miR-34b expression was positively correlated with disease-free survival and overall survival in breast cancer patients [8]. In gastric cancer, miR-34b expression was downregulated, and such effect was related to poor clinicopathological factors [9]. In prostate cancer, miR-34b expression was diminished in tumor tissues. Prostate cancer patients with high miR-34 expression presented longer overall and recurrence-free survival periods than those with low miR-34b levels [10]. In pancreatic cancer, miR-34b expression levels were lower in the tumor tissues and cell lines. Low miR-34b expression was significantly correlated with TNM stage, lymph-node metastasis and overall survival [11]. However, miR-34b expression was upregulated in colon cancer tissues. High miR-34b expression was strongly correlated with poor cancer-specific mortality [12]. These findings suggest that miR-34b expression exhibits tissue specificity and may present as a new promising prognostic biomarker in the abovementioned human cancer types. In OS, miR-34b expression was found to be decreased in the plasma and OS tissues. However, the biological roles and underlying regulatory mechanism of miR-34b in OS remains poorly understood. In this study, we focused on the biological roles of miR-34b in OS and determined the underlying regulatory mechanism. We found that miR-34b expression was downregulated in OS tissues and cell lines. Resumption of miR-34b expression hindered the proliferation and invasion of OS. Additionally, CCND1 was identified as a direct target of miR-34b in OS.
Material and Methods
Tissue samples and cell lines
A total of 26 paired OS tissues and corresponding adjacent normal tissues were collected from patients who underwent surgery at Fourth Hospital of Hebei Medical University. No chemotherapy or radiotherapy was administered to these OS patients prior to surgical resection. This research was approved by the ethics committee of the Fourth Hospital of Hebei Medical University. In addition, written informed consent was provided by all the patients.
Three OS cell lines (MG-63, U2OS and Saos-2) and the human normal osteoblasts (hFOB1.19) were purchased from American Type Culture Collection (Manassas, VA, USA). All cell lines were grown in RPMI-1640 medium containing 10% Foetal Bovine Serum (FBS), 100 U/ml penicillin and 100 μg/ml streptomycin (all from Gibco, Grand Island, NY, USA) and then cultured at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Cell transfection
MiR-34b mimics and miRNA negative control mimics (miRNC) were chemically synthesised by GenePharma Co., Ltd. (Shanghai, China). Cell transfection was carried out using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer’s instructions. The transfection mixture was replaced with fresh RPMI-1640 medium containing 10% FBS at 8 h posttransfection.
RNA isolation and quantitative reverse-transcription PCR (RT-qPCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was used to isolate total RNA from tissues or cells. MiR-34b levels were detected by reverse transcription through a TaqMan MiRNA Reverse-transcription kit (Applied Biosystems, Carlsbad, CA, USA) followed by quantitative PCR (qPCR) with a TaqMan MiRNA PCR Kit (Applied Biosystems, Carlsbad, CA, USA). Meanwhile, the mRNA levels of CCND1 were quantified using the Moloney murine leukaemia virus reverse transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) to synthesise cDNA. qPCR was performed using a SYBR-Green Master Mix kit (Roche Applied Science, Shanghai, China). U6 and Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) were used as normalisation controls for miR-34b and CCND1 mRNA expression, respectively. Data were analysed by the 2-ΔΔCq method.
Cell counting kit 8 (CCK8) assay
CCK8 assay was performed to detect cell proliferation. Transfected cells were collected at 24 h post-transfection and seeded onto 96-well plates at a density of 3 × 103 cells per well. Four time points, 0, 24, 48 and 72 h, were selected, and CCK8 assay was performed at each time point. Briefly, 10 μL of CCK8 solution (Dojindo Molecular Technologies, Kumamoto, Japan) was added to each well and incubated at 37°C for another 2 h. The absorbance of each well was determined at a wavelength of 450 nm by using a multi-well plate reader.
Transwell invasion assay
Matrigel-coated Transwell chambers with 8 μm pores (Coring, Inc., Corning, NY, USA) were employed to determine the cell invasion capacity. Briefly, 300 μL FBS-free cell suspension containing 5 × 104 cells was plated onto the upper chambers. Meanwhile, the lower chambers were filled with 500 μL of RPMI-1640 medium with 10% FBS. After 24 h of incubation, the cells that had not invaded the membrane in the chambers were gently removed using cotton swabs. In contrast, the invasive cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Cells five random fields for each Transwell chamber were counted under an inverted microscope.
Luciferase reporter assay
Luciferase reporter plasmids, pGL3-CCND1-3’-UTR Wild Type (WT) and pGL3-CCND1-3’-UTR Mutant (MUT), were synthesized and confirmed by GenePharma Co., Ltd. Cells were seeded into 24-well plates with a density of 60%-70% confluence. MiR-34b mimics or miR-NC was introduced into cells, together with pGL3-CCND1-3’-UTR WT or pGL3-CCND1-3’-UTR Mut using Lipofectamine 2000. Following transfection for 48 h, the luciferase activities were examined by the Dual-Luciferase Reporter Assay System (Promega Corp., Madison, WI, USA). The firefly luciferase activity was used as a normalization control for renilla luciferase activity.
Western blot analysis
Total protein was extracted using a RIPA lysis buffer (Beyotime Institute of Biotechnology, Nantong, China). A BCA kit (Beyotime Institute of Biotechnology) was adopted to measure the total protein concentration. Equal amounts of total protein was separated in 10% sodium dodecyl sulphate denatured polyacrylamide gel and transferred onto polyvinylidene fluoride membranes. After blocking in 5% non-fat milk for 1 h, the membranes were incubated overnight at 4°C with primary antibodies against CCND1 (sc-450) and GAPDH (sc-47724; all from Santa Cruz Biotechnology, CA, USA). Afterwards, the membranes were probed with goat anti-mouse peroxidase-conjugated secondary antibody (sc-2005; Santa Cruz Biotechnology, CA, USA) at room temperature for 2 h. Finally, an enhanced chemiluminescent reagent was utilised to visual the protein bands.
Statistical analysis
All data are shown as means ± standard deviations and analysed with student’s t-test or one-way ANOVA. SPSS 16.0 (SPSS, Chicago, IL, USA) was used for statistical analysis. P<0.05 indicated statistically significant differences.
Results
Expression and functions of miR-34b in OS
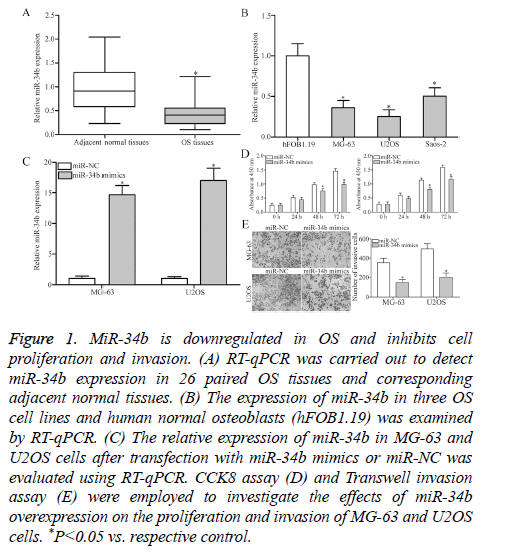
To investigate the exact roles of miR-34b in OS, RT-qPCR was performed to detect miR-34b expression in 26 paired OS tissues and corresponding adjacent normal tissues. MiR-34b expression was obviously downregulated in OS tissues than that in adjacent normal tissues (Figure 1A, P<0.05). Additionally, the expression levels of miR-34b in three OS cell lines (MG-63, U2OS and Saos-2) and human normal osteoblasts (hFOB1.19) were examined. RT-qPCR analysis results revealed that the miR-34b expression levels were lower in all the examined OS cell lines than in hFOB1.19 (Figure 1B, P<0.05). The potential roles of miR-34b in OS progression was then determined by transfecting MG-63 and U2OS cells with miR-34b mimics to increase the miRNA’s endogenous expression levels. RT-qPCR analysis confirmed that miR-34b was markedly upregulated in MG-63 and U2OS cells after transfection with miR-34b mimics (Figure 1C, P<0.05). Subsequently, the roles of miR-34b overexpression in regulating OS cell proliferation and invasion were evaluated by CCK8 assay and Transwell invasion assay. Cell proliferation (Figure 1D, P<0.05) and invasion (Figure 1E, P<0.05) were significantly decreased in the MG-63 and U2OS cells after miR-34b overexpression compared with those in the miR-NC group. These results suggest that miR-34b plays a tumor-suppressive role in OS.
Figure 1: MiR-34b is downregulated in OS and inhibits cell proliferation and invasion. (A) RT-qPCR was carried out to detect miR-34b expression in 26 paired OS tissues and corresponding adjacent normal tissues. (B) The expression of miR-34b in three OS cell lines and human normal osteoblasts (hFOB1.19) was examined by RT-qPCR. (C) The relative expression of miR-34b in MG-63 and U2OS cells after transfection with miR-34b mimics or miR-NC was evaluated using RT-qPCR. CCK8 assay (D) and Transwell invasion assay (E) were employed to investigate the effects of miR-34b overexpression on the proliferation and invasion of MG-63 and U2OS cells. *P<0.05 vs. respective control.
CCND1 is a direct target gene of miR-34b in OS
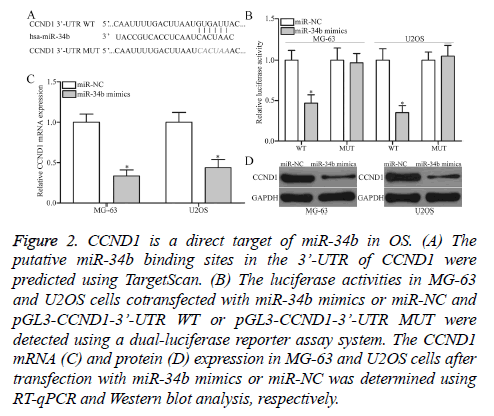
The mechanism underlying the tumor-suppressive roles of miR-34b in OS was determined by predicting the putative targets of miR-34b by TargetScan. CCND1 (Figure 2A) was predicated as a major target of miR-34b and selected for further confirmation. For confirmation, luciferase reporter assays were conducted in MG-63 and U2OS cells cotransfected with miR-34b mimics or miR-NC and pGL3-CCND1-3’-UTR WT or pGL3-CCND1-3’-UTR MUT. MiR-34b upregulation led to a significant decrease in the luciferase activities of the WT group in MG-63 and U2OS cells (P<0.05), but not in the MUT group, when compared with the miR-NC group (Figure 2B). Furthermore, the regulatory effect of miR-34b on endogenous CCND1 production was further assessed in MG-63 and U2OS. RT-qPCR and Western blot analysis data revealed that the ectopic expression of miR-34b reduced CCND1 expression in MG-63 and U2OS cells at the mRNA (Figure 2C, P<0.05) and protein (Figure 2D, P<0.05) levels. Overall, CCND1 is confirmed as a direct target of miR-34b in OS.
Figure 2: CCND1 is a direct target of miR-34b in OS. (A) The putative miR-34b binding sites in the 3’-UTR of CCND1 were predicted using TargetScan. (B) The luciferase activities in MG-63 and U2OS cells cotransfected with miR-34b mimics or miR-NC and pGL3-CCND1-3’-UTR WT or pGL3-CCND1-3’-UTR MUT were detected using a dual-luciferase reporter assay system. The CCND1 mRNA (C) and protein (D) expression in MG-63 and U2OS cells after transfection with miR-34b mimics or miR-NC was determined using RT-qPCR and Western blot analysis, respectively.
Discussion
MiRNAs deregulation has been reported to play important roles in tumorigenesis and tumor development through the regulation of their targets in OS. Hence, fully understanding the biological roles and mechanisms of OS-related miRNAs may contribute to the development of novel therapeutic targets for patients with this disease. In our current study, miR-34b expression levels were frequently reduced in OS tissues and cell lines. The resumption of miR-34b expression decreased cell proliferation and invasion in OS. Additionally, CCND1 was confirmed to be a direct target of miR-34b in OS. Overall, these findings suggest that miR-34b plays tumor-suppressive roles in OS by regulating CCND1.
MiR-34b has been implicated in the initiation and progression of human cancers. For instance, miR-34b upregulation led to a significant reduction in breast cancer growth in vitro and in vivo [13]. Majid et al. reported that the restoration of miR-34b expression suppressed cell growth and metastasis and promoted cell-cycle arrest and apoptosis in prostate cancer [10]. Liu et al. showed that the ectopic expression of miR-34b repressed pancreatic cancer cell migration and invasion in vitro and in vivo [11]. Dong et al. found that enforced expression of miR-34b in uveal melanoma inhibited cell growth and migration and induced cell cycle G [1] arrest [14]. Xiao et al. revealed that miR-34b re-expression impeded the cell viability and proliferation of nasopharyngeal carcinoma [15]. Li et al. discovered that miR-34b overexpression attenuated cell viability and increased cell apoptosis in acute myeloid leukaemia [16]. These findings suggest that miR-34b may be developed as a therapeutic target for treating such specific human cancer types.
Several targets of miR-34b have been validated. These targets include AKT [10] in prostate cancer, USP22 [15] in nasopharyngeal carcinoma, HSF1 [16] in acute myeloid leukaemia, SMAD3 [11] in pancreatic cancer and c-Met [14] in uveal melanoma. In the present study, CCND1 was a novel target of miR-34b in OS. CCND1, located on chromosome 11q13, is a well-known oncogene frequently highly expressed in multiple human cancer types. CCND1 is also overexpressed in OS tissues [17]. OS patients with high CCND1 expression presented with shorter overall survival periods than those of patients with low CCND1 expression [18]. CCND1 contributed to OS initiation and progression by helping regulate cell proliferation, the cell cycle, cell migration and cell invasion in vitro as well as growth and metastasis in vivo [18-20]. Considering the importance of CCND1 in OS, targeting CCND1 may be an effective therapeutic strategy for inhibiting the rapid tumor growth and metastasis in patients with OS.
In conclusion, the strengths of this study are that miR-34b inhibits OS cell proliferation and invasion by directly targeting CCND1. The study findings may provide a theoretical basis for the miRNA’s application in treating patients with OS. However, in this study, we did not explore the effect of miR-34b on OS cell migration, cell cycle, apoptosis in vitro and tumor growth in vivo. In our further experiments, we will investigate the roles of miR-34b in OS cell migration, cell cycle, apoptosis in vitro and tumor growth in vivo; as well as the upstream regulation mechanism.
References
- Maximov VV, Aqeilan RI. Genetic factors conferring metastasis in osteosarcoma. Future Oncol 2016; 12: 1623-1644.
- Pritchard DJ, Finkel MP, Reilly CA. The etiology of osteosarcoma. A review of current considerations. Clin Orthop Relat Res 1975; 14-22.
- Wang M, Xie R, Si H, Shen B. Integrated bioinformatics analysis of miRNA expression in osteosarcoma. Artif Cells Nanomed Biotechnol 2017; 45: 936-943.
- Moss EG. MicroRNAs: hidden in the genome. Curr Biol 2002; 12: 138-140.
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6: 857-866.
- Malan-Muller S, Hemmings SM, Seedat S. Big effects of small RNAs: a review of microRNAs in anxiety. Mol Neurobiol 2013; 47: 726-739.
- Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin 2014; 64: 311-336.
- Svoboda M, Sana J, Redova M. MiR-34b is associated with clinical outcome in triple-negative breast cancer patients. Diagn Pathol 2012; 7: 31.
- Tsai KW, Wu CW, Hu LY. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer 2011; 129: 2600-2610.
- Majid S, Dar AA, Saini S. miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin Cancer Res 2013; 19: 73-84.
- Liu C, Cheng H, Shi S. MicroRNA-34b inhibits pancreatic cancer metastasis through repressing Smad3. Curr Mol Med 2013; 13: 467-478.
- Hiyoshi Y, Schetter AJ, Okayama H. Increased microRNA-34b and -34c predominantly expressed in stromal tissues is associated with poor prognosis in human colon cancer. PLoS One 2015; 10: 0124899.
- Lee YM, Lee JY, Ho CC. miRNA-34b as a tumor suppressor in estrogen-dependent growth of breast cancer cells. Breast Cancer Res 2011; 13: 116.
- Dong F, Lou D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol Vis 2012; 18: 537-546.
- Xiao J, Li Y, Zhang W, Jiang Y, Du B, Tan Y. miR-34b inhibits nasopharyngeal carcinoma cell proliferation by targeting ubiquitin-specific peptidase 22. Onco Targets Ther 2016; 9: 1525-1534.
- Li G, Song Y, Zhang Y, Wang H, Xie J. miR-34b Targets HSF1 to Suppress Cell Survival in Acute Myeloid Leukemia. Oncol Res 2016; 24: 109-116.
- Wu J, Cui LL, Yuan J, Wang Y, Song S. Clinical significance of the phosphorylation of MAPK and protein expression of cyclin D1 in human osteosarcoma tissues. Mol Med Rep 2017; 15: 2303-2307.
- Han K, Chen X, Bian N. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget 2015; 6: 8875-8889.
- Ding Y, Fan DG, Shan LQ, Wang YC, Yang TT, Ma BA. ShRNA of Cyclin D1 decreased the proliferation of human osteosarcoma cell line SOSP-9607. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2009; 25: 1155-1157.
- Xu H, Mei Q, Shi L, Lu J, Zhao J, Fu Q. Tumor-suppressing effects of miR451 in human osteosarcoma. Cell Biochem Biophys 2014; 69: 163-168.