Research Article - Biomedical Research (2016) Health Science and Bio Convergence Technology: Edition-I
Leukemogenesis associated miRNAs regulate OSKM and Tp53 genes
Ehsan Valiollahi1, Javad Behravan1,2*1Biotechnology Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
2Department of Pharmaceutical Biotechnology, School of Pharmacy, Mashhad University of Medical Sciences, Mashhad, Iran
- *Corresponding Author:
- Javad Behravan
Biotechnology Research Center
Department of Pharmaceutical Biotechnology
School of Pharmacy
Mashhad University of Medical Sciences
Iran
Accepted date: May 25, 2016
Abstract
The last decade has witnessed an explosion of knowledge on microRNAs (miRNAs) and their functions in both normal and abnormal physiological contexts. Myeloid leukaemias are extremely diverse and complex diseases. Their occurrence has been demonstrated to be correlated with miRNAs expression aberrations. In order to identify miRNAs that have main role in regulation of OSKM factors and Tp53, here we present an in-depth analysis of miRNomes in human Acute Myeloid Leukaemia (AML) cell line, HL-60 and one human Chronic Myeloid Leukaemia (CML) cell line, K562, via high-throughput sequencing. Small RNA sequences obtained by high throughput sequencing of normal peripheral blood samples and myeloid cell lines K562 and HL60 were analysed to identify known miRNAs expression patterns. Target genes of the miRNAs were also identified using miRTarBase database. All the unique transcripts were used for the functional annotation exploiting the blast2go functional annotation tool. Our miRNome analysis identified about 50 miRNAs with complementary sites within target mRNAs of OSKM and Tp53 transcription factors. They included miR-335, miR-126, miR-34c, miR-21, miR-340, miR-10b, miR-25, miR-130a, miR-103a and miR-107. They were differentially expressed in myeloid cell lines. The unique expression patterns of miRNAs in cell lines confirm cell type and context dependent expression of miRNAs. Moreover, we identified a number of miRNAs that were differentially expressed in myeloid cell lines acting as regulators of OSKM and Tp53 transcription factor genes. These findings suggest the existence of complicated crosstalk mechanisms between identified differentially expressed miRNAs and these factors. This underlines the importance and complexity of the functional roles of dysregulated miRNAs in leukemogenesis. That act by disturbing some essential networks related to proliferation, differentiation, apoptosis, signal transduction, cell division and tumor suppression.
Keywords
MicroRNAs, Leukaemia, OSKM (OCT4, SOX2, KLF4 and MYC), Tp53.
Introduction
Myeloid leukaemias, including Acute Myeloid Leukaemia (AML) and Chronic Myeloid Leukaemia (CML), are clonal bone marrow diseases identified by the pathological proliferation of abnormal white blood cells [1]. It is assumed that Cancer Stem Cells (CSCs) originate from stem cells carrying mutations or dedifferentiated mature cells [2]. As stem cells possess longer life span compared to their progeny, it is supposed that stem cells have the highest potential to accumulate a mandatory number of mutations to disturb intrinsic mechanisms tuning normal cell metabolisms and proliferation. Furthermore, the dedifferentiation of mature cells possibly occurs in tumorigenesis. For instance, in the process of Epithelial Mesenchymal Transition (EMT), mature cell becomes more “stem cell-like” with specific dysregulated transcription factor genes, e.g. Sox2 and Klf4 [3]. Tp53 mutations are the most common genetic alteration associated with stem cell-like gene expression signatures in Hepatocellular Carcinoma (HCC) [4], and loss of p53 function enhances the efficiency of OSKM factors-induced reprogramming [5,6]. These data together propose that factors tune cellular reprogramming and may also be involved in CSCs generation as oncogenes during cell dedifferentiation process. In a similar context, Leukaemia Stem Cells (LSCs) are very similar to their normal counterparts, Haematopoietic Stem Cells (HSCs). These similarities are obvious in their function, surface antigenic properties, cytologic properties and gene expression patterns. Therefore, it has been hypothesized that AML and CML may arise as a result of accumulation of genetic and epigenetic events (including miRNA dysregulation) in HSCs [7]. In fact Yamanaka and colleagues demonstrated that differentiated cells can be reprogrammed to acquire pluripotency by specific transcription factors including Oct3/4, Sox2, Klf4, and Myc (Yamanaka or OSKM factors). These factors have been revealed to be activated in Embryonic Stem cells (ES cells) [8,9]. Likewise aberrant acquisition of stem-cell like characteristics through molecular mechanisms including miRNA dysregulation affecting the expression of OKSM genes may be a cause of transition from leukaemia cells to LSCs with self-renewing capabilities. The miRNA molecules are now known to play diverse regulatory functions in both plants and animals [10,11]. They are generally 19-24 nucleotides in length, small non-coding RNAs which can inhibit the translation or lead to degradation of mRNA, usually by binding to the 3/-Untranslated Regions (UTRs) of target mRNAs [12]. A set of miRNA-coding genes are closely distributed in genome and these closely organized miRNAs are termed as the miRNA cluster.
The role of miRNAs in mouse and human embryonic stem cells has been previously explored using Dicer1- and DGCR8- deficient cells, which are essential for miRNA biogenesis. Deletion of Dicer1 causes embryonic lethality in mice [13] and DGCR8-deficient mouse ESCs exhibit altered regulation of the cell cycle and differentiation that are associated with decline in silence stemness markers, such as Oct4, Rex1, Sox2, and Nanog, likewise delayed expression of differentiation markers [14].
As a repercussion, miRNA dysregulation could lead to disruption of the hematopoietic system and leukaemia may arise. Accumulating evidence implies that miRNAs have crucial functions in myeloid development and leukemogenesis. MiRNA expression profiling is gaining popularity because miRNA signatures have been associated with the diagnosis and prognosis of diseases such as leukaemia [15,16]. Here we present an in-depth analysis of miRNomes in human acute Myeloid Leukaemia (AML) cell line, HL-60 and one human Chronic Myeloid Leukaemia (CML) cell line, K562, via highthroughput sequencing to identify miRNAs that have main role in regulation of OSKM factors and Tp53. This could shed light on better understanding of the important aspects of molecular mechanisms of leukemogenesis.
Materials and Methods
sRNA libraries and sRNA-seq analysis
SRNA-seq data of normal blood samples, K562 and HL-60 leukaemia cell lines were obtained from the NCBI SRA (Sequence Read Archive). In brief samples were sequenced according to the following steps; total RNA was isolated from K562, HL-60 and normal peripheral blood cells using TRIZOL® Reagent. Total RNA was separated on denaturing polyacrylamide gel electrophoresis, then approximately 18-30 nucleotide RNAs were excised from and ligated to sequencing adaptors on the both ends, and were reversely transcribed using a sRNA sample prep kit (Illumina, San Diego, CA, USA) and then PCR amplified with adapter specific primers. The amplified cDNAs were finally purified on acrylamide gel to generate cDNA tag libraries for sequencing on the illumina platform. Sequences were then queried against ribosomal and transfer RNAs from Rfam (http://www.sanger.ac.uk/Software/Rfam/), the genomic tRNA database (http://gtrnadb.ucsc.edu/) and NCBI database (www.ncbi.nlm.nih.gov/) via Bowtie aligner [17]. Unmatched sequences retrieved for identify known miRNAs expression in the leukaemia cell lines and normal blood samples.
Target prediction and functional annotation
Target genes of miRNAs were identified using miRTarBase database, an information resource for experimentally validated miRNA-target interactions [18]. All the unique transcripts were taken for the functional annotation using the blast2go functional annotation tool [19].
Statistical analysis
To compare abundance of miRNAs in normal blood samples and leukaemia cell line libraries, the count of each miRNA was normalized to Transcripts per Million (TPM). The significance of differences in miRNA frequency between normal blood samples and leukaemia cell lines was calculated using a χ2 test. The thresholds we considered to identify differentially expressed miRNAs were fold-change ≥ 1.5 and p ≤ 0.05.
Results and Discussion
OSKM and Tp53 transcription factors and miRNAs as regulators
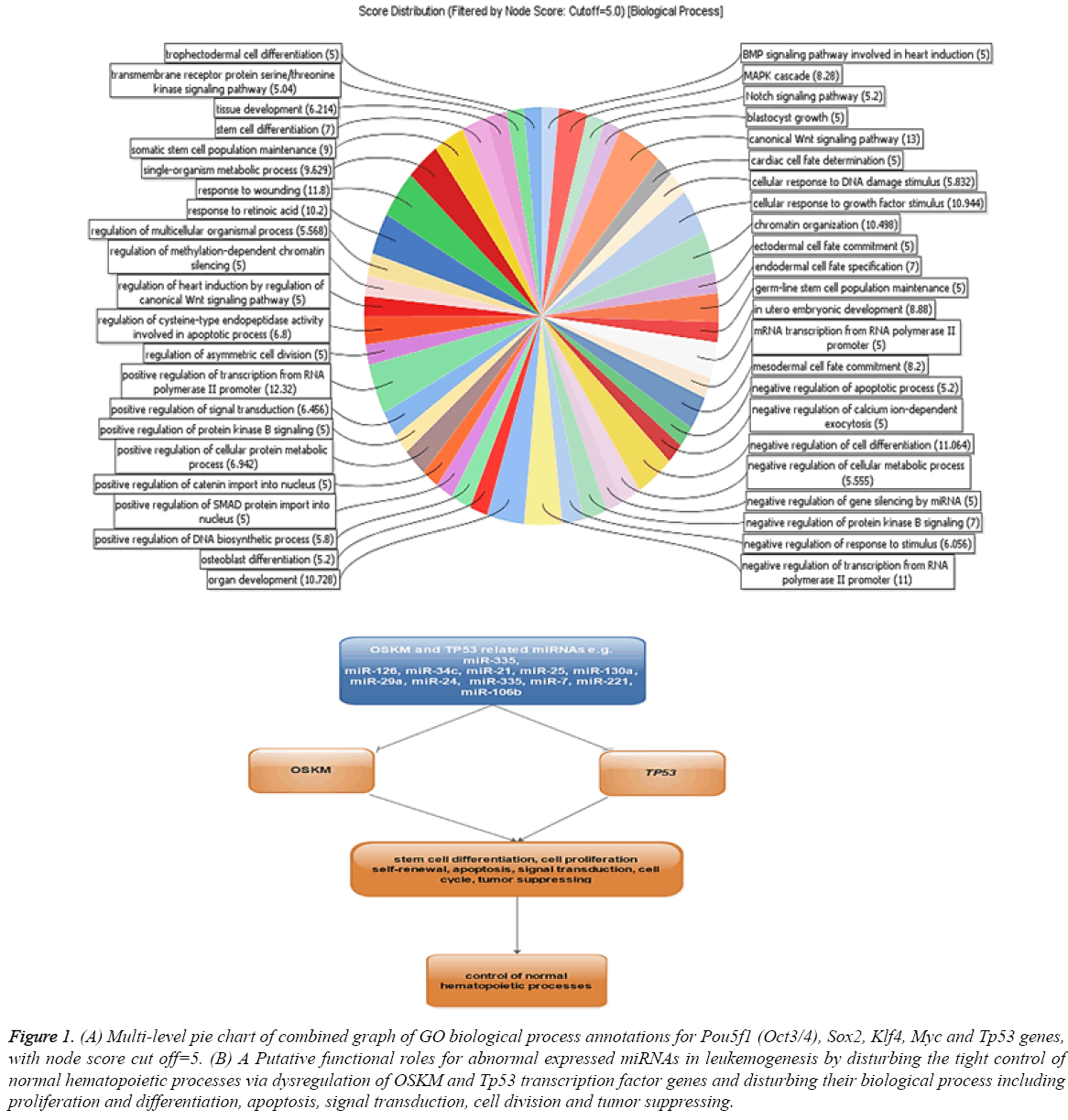
The dysregulation of some OSKM factors genes including Oct4, Sox2 and Klf4 in breast, ovarian cancer cells and leukaemia cells were reported [20-22]. A direct analysis about miRNAs function in tuning of these genes in myeloid cell lines has not been established. To determine this mechanism, we analysed the expression of known miRNAs in our selected cell lines and their interaction with OSKM and Tp53 transcription factor genes. To identify the miRNA interacting with OSKM and Tp53 transcription factor genes in leukaemia, the miRTarBase database with strong evidence criteria was used. Based on the obtained data about 4, 14, 15, 44 and 35 miRNAs with strong evidence were identified as regulators of Pou5f1 (Oct3/4), Sox2, Klf4, Myc and Tp53 genes, respectively (Table 1). Gene ontology analysis of putative target genes by Blast2Go revealed that the target genes were involved in some regulatory functions and biological processes including regulation of stem cell differentiation, apoptosis, signal transduction, cell division and stress response (Figure 1). These biological processes may imply the functions of OSKM and p53 transcription factors in induction of tumors. Dysregulation of Sox2 promotes the tumorigenesis process and increases apoptosis-resistance properties in prostate cancer cells [23]. This study exemplifies that miRNAs may strongly contribute to leukemogenesis by disturbing the tight control of normal hematopoietic processes via dysregulation of OSKM and Tp53 transcription factor genes (Figure 1).
| Target genes | Identified miRNAs with strong evidence |
|---|---|
| Pou5f1 | miR-145-5p, miR-128-3p, miR-335-5p, miR-34a-5p |
| Sox2 | miR-145-5p, miR-126-3p, miR-522-3p, miR-34a-5p, miR-34b-3p, miR-34c-5p, miR-140-5p, miR-429, miR-128-3p, miR-21-5p, miR-340-5p, miR-625-5p, miR-638, miR-1181 |
| Klf4 | miR-145-5p, miR-10b-5p, miR-25-3p, miR-135b-5p, miR-130a-3p, miR-663a, miR-103a-3p, miR-107, miR-128-3p, miR-124-3p, miR-29a-3p, miR-15a-5p, miR-34a-5p, miR-7-5p, miR-137, miR-152-3p, miR-206 |
| Myc | miR-24-3p, let-7a-5p, let-7g-5p, miR-335-5p, let-7f-5p, miR-744-5p, miR-148a-3p, miR-212-3p, miR-494-3p, miR-155-5p, miR-33a-5p, miR-487b-3p, miR-7-5p, miR-93-5p, miR-324-3p, miR-184, miR-126-5p, miR-25-3p, miR-92a-2-5p, miR-92a-1-5p, miR-19b-2-5p, miR-19b-1-5p, miR-19a-3p, miR-106b-5p, miR-34a-5p, miR-98-5p, let-7c-5p, miR-26a-5p, miR-145-5p, miR-21-5p, miR-34b-5p, miR-34c-5p, miR-18a-5p, miR-17-5p, miR-20a-5p, miR-34b-3p, miR-378a-3p, miR-371a-3p, miR-373-3p, miR-451a, miR-33b-5p, miR-135a-5p, miR-449c-5p, miR-429 |
| Tp53 | miR-125b-5p, miR-125a-5p, miR-25-3p, miR-30d-5p, miR-1285-3p, miR-612, miR-15a-5p, miR-16-5p, miR-221-3p, miR-222-3p, miR-214-3p, miR-10b-5p, miR-608, miR-605-5p, miR-504-5p, miR-485-5p, miR-27a-3p, miR-150-5p, miR-92a-3p, miR-375, miR-200a-3p, miR-491-5p, miR-30a-5p, miR-30b-5p, miR-30c-5p, miR-30e-5p, miR-19b-3p, miR-92a-2-5p, miR-92a-1-5p, miR-20a-5p, miR-18a-5p, miR-106b-5p, miR-106a-5p, miR-17-5p |
Table 1: Identified miRNAs with complementary sites within mRNAs of Pou5f1, Sox2, Klf4, Myc and Tp53 genes according strong evidence in miRTarBase database.
Figure 1: (A) Multi-level pie chart of combined graph of GO biological process annotations for Pou5f1 (Oct3/4), Sox2, Klf4, Myc and Tp53 genes,with node score cut off=5. (B) A Putative functional roles for abnormal expressed miRNAs in leukemogenesis by disturbing the tight control of normal hematopoietic processes via dysregulation of OSKM and Tp53 transcription factor genes and disturbing their biological process including proliferation and differentiation, apoptosis, signal transduction, cell division and tumor suppressing.
Expression of miRNAs in CML and AML cell lines
The representative cell lines were obtained from patients with myeloid leukaemia and therefore can represent clinical aspects of myeloid leukaemia to some extent. To explore the underlying mechanism of miRNA expression and the relationship between the samples, collected sequences were perfectly aligned to known human miRNAs. According to our results some of the identified miRNAs with complementary sites within target mRNAs of Pou5f1, Sox2, Klf4, Myc and Tp53 genes, were not expressed in normal peripheral blood samples and leukaemia cell lines. This pattern may be associated with miRNAs tissue specificity, indicating that miRNAs expression may be regulated both temporally and spatially [24,25]. To demonstrate miRNA function in leukemogenesis and identify miRNAs which are dysregulated in acute leukaemia, genome-wide analysis of miRNA expression studies have been carried out. These works revealed that leukemic cells of acute leukaemia patients had dissimilar miRNA expression patterns compared to normal hematopoietic cells [15,26-28].
Our miRNome analysis confirmed these results and about 50 miRNAs with complementary sites within target mRNAs of OSKM and Tp53 transcription factors, including miR-335, miR-126, miR-34c, miR-21, miR-340, miR-10b, miR-25, miR-130a, miR-103a and miR-107 were differentially expressed in myeloid cell lines. Some of the differentially expressed miRNAs such as miR-126, miR-34c, miR-340, miR-130a, miR-335 and miR-7 were down regulated in both cell lines (Table 2). This pattern may be considered as a common function of these miRNAs in leukemogenesis. About 50 miRNAs were differentially expressed in myeloid cell lines, whereas in HL60 miR-124, miR-487b, miR-1285, miR-504, miR-485 and in K562 miR-128, miR-10b, miR-92a, miR-30d, miR-375 were uniquely expressed. These patterns confirm cell type and context dependent expression of miRNAs (Supplementary File 1). Analysis of Pou5f1-linked miRNAs revealed miR-128 and miR-335 as regulator for Pou5f1 in HL60 and K562 cell lines and miR-335 were down regulated in both cell lines (Tables 2 and 3). Aberrant expression of this gene has been reported in several human and rat tumor cells [29,30]. All expressed miRNAs in HL60 and K562 cell lines with target sites within Sox2 mRNA, were down regulated in both cell lines (Table 2). This may indicate common function of these miRNAs in leukemogenesis via dysregulation of Sox2 and may cause increased expression of this gene in AML and CML cell lines. Aberrant expression of Sox2 has been reported in leukaemia and a number of solid tumors, including cancers of the prostate and brain. This may be correlated with increased proliferation and tumorigenicity of cancer stem cells [31-34].
| miRNAs | Target genes | Expression HL60 | Expression K562 | miRNAs | Target genes | Expression Hl60 | Expression K562 |
|---|---|---|---|---|---|---|---|
| miR-128-3p | Pou5f1 | N | Down | miR-155-5p | Myc | Up | Down |
| miR-335-5p | Pou5f1 | Down | Down | miR-487b-3p | Myc | Down | N |
| miR-126-3p | Sox2 | Down | Down | miR-7-5p | Myc | Down | Down |
| miR-34c-5p | Sox2 | Down | Down | miR-93-5p | Myc | Up | Down |
| miR-128-3p | Sox2 | N | Down | miR-126-5p | Myc | N | Up |
| miR-21-5p | Sox2 | Down | Down | miR-25-3p | Myc | Up | Up |
| miR-340-5p | Sox2 | Down | Down | miR-92a-1-5p | Myc | N | Up |
| miR-10b-5p | Klf4 | N | Up | miR-19b-1-5p | Myc | Up | Up |
| miR-25-3p | Klf4 | Down | Up | miR-19a-3p | Myc | Down | Up |
| miR-130a-3p | Klf4 | Down | Down | miR-106b-5p | Myc | Down | Down |
| miR-103a-3p | Klf4 | Up | Down | miR-125b-5p | Tp53 | Down | Up |
| miR-107 | Klf4 | Up | Down | miR-125a-5p | Tp53 | Down | Down |
| miR-128-3p | Klf4 | N | Down | miR-25-3p | Tp53 | Down | Up |
| miR-124-3p | Klf4 | Up | N | miR-30d-5p | Tp53 | N | Down |
| miR-29a-3p | Klf4 | Down | Down | miR-1285-3p | Tp53 | Down | N |
| miR-15a-5p | Klf4 | Up | Down | miR-15a-5p | Tp53 | Up | Down |
| miR-7-5p | Klf4 | Down | Down | miR-16-5p | Tp53 | Up | Down |
| miR-152-3p | Klf4 | Up | Down | miR-221-3p | Tp53 | Up | Down |
| miR-24-3p | Myc | Down | Down | miR-222-3p | Tp53 | Up | Down |
| let-7a-5p | Myc | Up | Down | miR-10b-5p | Tp53 | N | Up |
| let-7g-5p | Myc | Down | Down | miR-504-5p | Tp53 | Down | N |
| miR-98-5p | Myc | Up | Down | miR-485-5p | Tp53 | Down | N |
| let-7c-5p | Myc | Up | Down | miR-27a-3p | Tp53 | Up | Down |
| miR-26a-5p | Myc | Down | Down | miR-150-5p | Tp53 | Down | Down |
| miR-21-5p | Myc | Down | Down | miR-92a-3p | Tp53 | Up | Up |
| miR-34c-5p | Myc | Down | Down | miR-375 | Tp53 | N | Down |
| miR-18a-5p | Myc | Down | Up | miR-30b-5p | Tp53 | Up | Up |
| miR-17-5p | Myc | Up | Down | miR-30c-5p | Tp53 | Up | Up |
| miR-20a-5p | Myc | Up | Up | miR-19b-3p | Tp53 | Down | Up |
| miR-378a-3p | Myc | Down | Down | miR-92a-1-5p | Tp53 | N | Up |
| miR-451a | Myc | Down | Down | miR-20a-5p | Tp53 | Up | Up |
| miR-335-5p | Myc | Down | Down | miR-18a-5p | Tp53 | Down | Up |
| let-7f-5p | Myc | Up | Down | miR-106b-5p | Tp53 | Down | Down |
| miR-744-5p | Myc | Down | Down | miR-17-5p | Tp53 | Up | Down |
| miR-148a-3p | Myc | Down | Down |
Table 2: Differentially expressed miRNAs related to OSKM and Tp53 in AML (HL60) and CML (K562) cell lines, compare to normal peripheral blood cells. N, indicate miRNA that are NOT expressed in AML (HL60) or CML (K562) cell lines.
| Duplex structure | Position | Score | MFE |
|---|---|---|---|
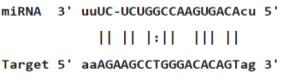 |
230-251 | 107 | -11.7 |
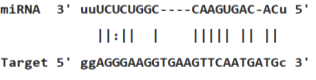 |
161-186 | 97 | -9.2 |
 |
88-108 | 90 | -5.5 |
Table 3: miR-128 binding sites in 3'UTR of Pou5f1 mRNA as regulator for Pou5f1 (predicted by miRanda). Minimum Free Energy (MFE) is calculated by miRanda.
Moreover, Sox2 up regulation has been reported in several epithelial tumors [34-37] and is indicative of the function of Sox2 in tumorigenesis of cancer cells. Xenotransplantation experiments in NSG mice showed that Sox2-overexpressing cells induced tumors earlier and more frequent than control serous ovarian carcinoma cells. Sox2 expression facilitating transition to a CSC state with enhanced tumor-initiating properties [33]. Furthermore, we identified some differentially expressed miRNAs between AML and CML cell lines that were associated with Klf4, Myc and Tp53 expression. For instance miR-10b, miR-25, miR-103a, miR-107 and miR-29a targeting Klf4 and miR-25, miR-15a, miR-16, miR-221, miR-222 and miR-27a as Tp53 regulators show contrary expression between AML and CML cell lines. MiR-16 was up regulated in AML whereas it was down regulated in CML cell lines significantly (Table 2). Abnormal expression of some miRNAs, such as miR-16, 151 and 142 were formerly reported in CML [38-40]. This cell specific pattern may be related to the physiological characteristics of these cell lines, CML and AML pathogenesis-related pathways and also genetic alterations in AML and Philadelphia chromosome (Ph) in CML cell lines [41-43]. Hematologic malignancies often display genomic alterations. Balanced and unbalanced chromosome alterations are established in AML patients [44].
Analysis of human and mouse genomes exhibited that most of miRNA genes were located at fragile sites and altered genomic regions that were involved in cancer [45]. These data imply that in part the mechanism of miRNA deregulation in leukemogenesis may be due to genomic instability. The unique miRNA expression pattern in AML and CML cell lines may be associated with cytogenetic aberrations and may reveal insights into the biology of leukaemia subtypes. MYC has a fundamental function in the correct haematopoiesis, tuning the delicate balance among self-renewal, differentiation, and proliferation essential for blood formation.
According to our data, most of miRNAs with complementary sites within Myc mRNA in CML cell line were down regulated (Table 2). Up regulation of c-Myc mRNA with CML progression has been reported before. MYC activity can elicit genomic instability and leukemognesis [46]. Down regulation of identified miRNAs may be related to up regulation of Myc in K562 cell line. This confirms that a role is played by these miRNAs in tuning Myc expression.
Myc deregulation is frequently exposed in different types of lymphoma and leukaemia, which would demolish the required equilibrium for blood formation and transform hematopoietic cells by triggering proliferation and blocking terminal differentiation [47]. P53 is a tumor suppressor and sequencespecific transcription factor that regulates expression of genes involved in apoptosis, cell cycle arrest and senescence. P53 can also inhibit expression of some genes. This function is partly mediated by inducing several miRNAs. Indeed, p53 is mutated in about 50% of human cancers, and is functionally inactivated in many more. The crosstalk between the Tp53 network and miRNAs is further substantiated by recent studies on regulation of Tp53 by miRNAs. Several miRNAs including miR-125b, miR-504, miR-25, miR-30d, miR-122, miR-29, miR-192, miR-194 and miR-215 have been indicated to regulate p53 abundance and activity. Among these miR-125b, miR-504, miR-25 and miR-30d negatively regulate Tp53 gene by binding to its 3’UTR (Table 4) [48,49]. Therefore, altered expression of miRNAs associated with these transcription factors in myeloid cell lines may have a critical function in leukemogenesis by disturbing biological processes regulated by these molecules.
| Duplex structure | Position | Score | MFE |
|---|---|---|---|
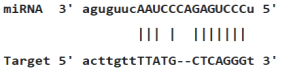 |
1010-1029 | 147.00 | -15.20 |
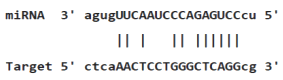 |
1268-1289 | 130.00 | -15.44 |
 |
1475-1494 | 124.00 | -18.00 |
Table 4: miR-125b binding sites in 3'UTR of Tp53 mRNA predicted by miRanda. Minimum Free Energy (MFE) is calculated by miRanda.
Conclusion
The discovery of target regulation by miRNAs may shed new lights on comprehension of leukemogenesis mechanisms. Myeloid leukaemias are extremely diverse diseases and have been demonstrated to be correlated with miRNAs expression aberrations. In this work, we identified a number of miRNAs that were differentially expressed in myeloid cell lines as regulators of OSKM and Tp53 transcription factor genes. This suggests the existence of crosstalk mechanisms between identified differentially expressed miRNAs and these factors which underlies the functional roles of dysregulated miRNAs in leukemogenesis via disturbing some essential networks related to proliferation, differentiation, apoptosis, signal transduction, cell division and tumor suppression (Figure 1). The unique miRNA expression patterns that were found for subtypes of myeloid cell lines may contribute to the biology of diverse leukaemia subtypes. Accordingly, we deduced that some of the uniquely expressed miRNAs between the AML and CML cell lines may discriminate between these two myeloid leukaemia types and act as diagnostic, predictive and prognostic biomarkers. This indicates that miRNAs are potentially useful in categorizing leukaemia subtypes and that specific classification may be useful for improvement clinical approach such as diagnosis and treatment strategies for myeloid leukaemia. This study provides useful data to further explore the mechanisms of microRNA-mediated gene regulation in human leukaemia subtyping, leukemogenesis and myeloid development.
References
- Murati A, Brecqueville M, Devillier R, Mozziconacci MJ, Gelsi-Boyer V. Myeloid malignancies: mutations, models and management. BMC Cancer 2012; 12: 304.
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW. Creation of human tumour cells with defined genetic elements. Nature 1999; 400: 464-468.
- Qi W, Liang W, Jiang H, Miuyee Waye M. The function of miRNA in hepatic cancer stem cell. Biomed Res Int 2013; 2013: 358902.
- Woo HG, Wang XW, Budhu A, Kim YH, Kwon SM. Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma. Gastroenterology 2011; 140: 1063-1070.
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 2009; 460: 1132-1135.
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009; 460: 1145-1148.
- Chung SS, Hu W, Park CY. The role of microRNAs in hematopoietic stem cell and leukemic stem cell function. Ther Adv Hematol 2011; 2: 317-334.
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131: 861-872.
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663-676.
- Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 2007; 210: 279-289.
- Valiollahi E, Farsi M, Mirshamsi A. Sly-Mir166 and Sly-Mir319 are components of the cold stress response in Solanum lycopersicum. Plant Biotechnol Reports 2014; 8: 349-356.
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010; 466: 835-840.
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H. Dicer is essential for mouse development. Nat Genet 2003; 35: 215-217.
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 2007; 39: 380-385.
- Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood 2008; 111: 3183-3189.
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukaemia from acute myeloid leukaemia. Proc Natl Acad Sci USA 2007; 104: 19971-19976.
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25.
- Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, Tsai TR, Ho SY, Jian TY, Wu HY, Chen PR, Lin NC, Huang HT, Yang TL, Pai CY, Tai CS, Chen WL, Huang CY, Liu CC, Weng SL, Liao KW, Hsu WL, Huang HD. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016; 44: D239-D247.
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005; 21: 3674-3676.
- Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012; 31: 1354-1365.
- Peng S, Maihle NJ, Huang Y. Pluripotency factors Lin28 and Oct4 identify a sub-population of stem cell-like cells in ovarian cancer. Oncogene 2010; 29: 2153-2159.
- Kharas MG, Yusuf I, Scarfone VM, Yang VW, Segre JA. KLF4 suppresses transformation of pre-B cells by ABL oncogenes. Blood 2007; 109: 747-755.
- Jia X, Li X, Xu Y, Zhang S, Mou W. SOX2 promotes tumorigenesis and increases the anti-apoptotic property of human prostate cancer cell. J Mol Cell Biol 2011; 3: 230-238.
- Cuellar TL, McManus MT. MicroRNAs and endocrine biology. J Endocrinol 2005; 187: 327-332.
- Krichevsky AM, Gabriely G. miR-21: A small multi-faceted RNA. J Cell Mol Med 2009; 13: 39-53.
- Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One 2008; 3: e2141.
- Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukaemia. Blood 2008; 111: 5078-5085.
- Li Z, Lu J, Sun M, Mi S, Zhang H. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA 2008; 105: 15535-15540.
- Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis 2005; 26: 495-502.
- Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene 2001; 20: 8085-8091.
- Gelebart P, Hegazy SA, Wang P, Bone KM, Anand M, Sharon D, Hitt M, Pearson JD, Ingham RJ, Ma Y, Lai R. Aberrant expression and biological significance of Sox2, an embryonic stem cell transcriptional factor, in ALK-positive anaplastic large cell lymphoma. Blood Cancer J 2012; 2: e82.
- Apostolou P, Toloudi M, Chatziioannou M, Ioannou E, Papasotiriou I. Cancer stem cells stemness transcription factors expression correlate with breast cancer disease stage. Curr Stem Cell Res Ther 2012; 7: 415-419.
- Phi JH, Park SH, Kim SK, Paek SH, Kim JH. Sox2 expression in brain tumors: a reflection of the neuroglial differentiation pathway. Am J Surg Pathol 2008; 32: 103-112.
- Sattler HP, Lensch R, Rohde V, Zimmer E, Meese E. Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate 2000; 45: 207-215.
- Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S. SOX2 is an amplified lineage-survival oncogene in lung and oesophageal squamous cell carcinomas. Nat Genet 2009; 41: 1238-1242.
- Hussenet T, Dali S, Exinger J, Monga B, Jost B. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS One 2010; 5: e8960.
- Zhang J, Chang DY, Mercado-Uribe I, Liu J. Sex-determining region Y box2 expression predicts poor prognosis in human ovarian carcinoma. Hum Pathol 2012; 43:1405-1412.
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834-838.
- Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, Bandres E, Cordeu L, Aparicio O, Saez B, Navarro G, Vilas-Zornoza A, Perez-Roger I, Garcia-Foncillas J, Torres A, Heiniger A, Calasanz MJ, Fortes P, Roman-Gomez J, Prosper F. Down-regulation of hsa-miR-10a in chronic myeloid leukaemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res 2008; 6: 1830-1840.
- Ramkissoon SH, Mainwaring LA, Ogasawara Y, Keyvanfar K, McCoy JP Jr. Hematopoietic-specific microRNA expression in human cells. Leuk Res 2006; 30: 643-647.
- Starczynowski DT, Morin R, McPherson A, Lam J, Chari R. Genome-wide identification of human microRNAs located in leukaemia-associated genomic alterations. Blood 2011; 117: 595-607.
- Daschkey S, Rottgers S, Giri A, Bradtke J, Teigler-Schlegel A, Meister G, Borkhardt A, Landgraf P. MicroRNAs distinguish cytogenetic subgroups in paediatric AML and contribute to complex regulatory networks in AML-relevant pathways. PLoS One 2013; 8: e56334.
- Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukaemia. Blood 2000; 96: 3343-3356.
- Marcucci G, Radmacher MD, Maharry K, Mrozek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med 2008; 358: 1919-1928.
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004; 101: 2999-3004.
- Albajar M, Gomez-Casares MT, Llorca J, Mauleon I, Vaque JP. MYC in chronic myeloid leukemia: induction of aberrant DNA synthesis and association with poor response to imatinib. Mol Cancer Res 2011; 9: 564-576.
- Delgado MD1, Leon J. Myc roles in haematopoiesis and leukaemia. Genes Cancer 2010; 1: 605-616.
- Sinnett D, Richer C, Deragon JM, Labuda D. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J Biol Chem 1991; 266: 8675-8678.
- Jones M, Lal A. MicroRNAs, wild-type and mutant p53: more questions than answers. RNA Biol 2012; 9: 781-791.