Mini Review - Asian Journal of Biomedical and Pharmaceutical Sciences (2023) Volume 13, Issue 101
Iron-Sulfur cluster and cancer related diseases
Mohammad Deylam Kamar*Department of chemistry, University of Isfahan, Iran
- *Corresponding Author:
- Mohammad Deylam Kamar
Department of chemistry
University of Isfahan, Iran
E-mail: mobindeylam@gmail.com
Received: 29-Aug -2023, Manuscript No. AABPS-23- 114709; Editor assigned: 01-Sep -2023, PreQC No. AABPS-23-114709(PQ); Reviewed: 14-Sep -2023, QC No. AABPS-23-114709; Revised: 20-Sep-2023, Manuscript No. AABPS-23-114709 (R); Published: 29-Sep -2023, DOI: 10.35841/ aabps-13.101.193
Citation: Deylam Kamar M. Iron-Sulfur cluster and cancer related diseases. Asian J Biomed Pharmaceut Sci. 2023;13(101):193
Abstract
Iron-sulfur (Fe-S) clusters are essential protein cofactors involved in various cellular processes. Defects in Fe-S cluster biogenesis can lead to diseases like cancer. The stability and transfer of Fe-S clusters mediated by proteins like NAF-1 regulate cellular resistance to oxidative stress. This study examined how modulating NAF-1 and Fe-S cluster stability affected breast cancer tumor growth in MDA-MB-231 cells. Overexpression of NAF-1 enhanced antioxidant capacity and oxidative stress resistance, increasing tumor volume 3-fold. In contrast, a stabilizing mutation in NAF-1 inhibited cluster transfer to recipient proteins, decreasing antioxidant capacity and reducing tumor volume by 30%. The drug pioglitazone had similar effects to the NAF-1 mutation, hyperstabilizing its Fe-S cluster and suppressing its function. High cancer cell metabolism produces reactive oxygen species that are normally detoxified. New cancer therapies exploit this by overloading cells with oxidative radicals using iron nano-catalysts. Thus, understanding factors regulating Fe-S cluster stability in cancer cells provides opportunities to enhance prooxidant cancer therapies. Overall, this study demonstrated that increased Fe-S cluster stability reduces NAF-1 mediated antioxidant capacity and cancer cell growth, while destabilization enhances sensitivity to oxidative stress. Modulating proteins involved in Fe-S cluster transfer represents a potential therapeutic approach for cancer treatment.
Introduction
Iron-Sulfur (Fe-S) clusters are ancient, versatile protein cofactors that are critical for various cellular processes including electron transfer, regulation of gene expression, and metabolism. Defects in Fe-S cluster biogenesis are linked to diseases like Friedreich's ataxia, sideroblastic anemia, and some cancers. The iron-sulphur cluster (ISC) assembly system in mitochondria is the primary machinery for Fe-S cluster synthesis in eukaryotes. The ISC components themselves contain Fe-S clusters, highlighting the importance of their proper maturation. The NEET family protein NAF-1 plays emerging roles in iron homeostasis, autophagy, cell death pathways, and regulating cellular resistance to oxidative stress. Cancer cells have adapted to handle high levels of reactive oxygen species (ROS) from their elevated metabolism. New cancer therapies use iron nano-catalysts to overload cancer cells with oxidative radicals. Therefore, understanding factors that regulate Fe-S cluster stability and transfer in cancer cells may reveal opportunities to enhance these pro-oxidant cancer therapies. This study examined how modulating the stability of Fe-S clusters mediated by NAF-1 affects antioxidant capacity and breast cancer tumor growth.
History of Iron-Sulfur clusters
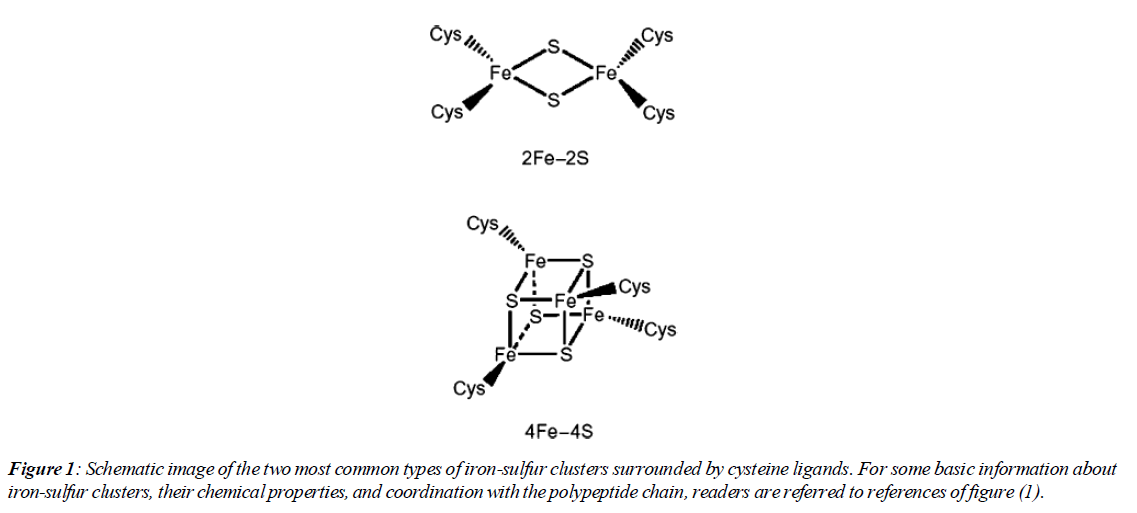
The existence of iron-sulfur cluster proteins was investigated through various sources such as photosynthetic organisms, nitrogen-fixing bacteria, mammalian mitochondrial codons. Initially in the 1960s, information about them was at the level of a group of unknown proteins that contained iron atoms and had a role in redox functions. In the mid-1960s, it was revealed that these proteins contain a complex of iron and cysteine sulfur atoms [1].More data on their electronic and geometric structure was obtained later and it was shown that they are analogs of the active site of proteins. The result of these findings showed that the active site does not necessarily have to have a protein structure (Figure 1).
Methods
MDA-MB-231 human breast cancer cells with NAF- 1 (Control), suppressed NAF-1[shRNA; NAF-1(-)], or overexpressed NAF-1[NAF-1(+) or NAF-1(H114C)] were injected into female athymic node 5- to 6-wk-old mice. Also, mouse weight and tumor size were measures twice weekly throughout the experiment.
Examination of assembly and construction of clusters
In Prokaryotes
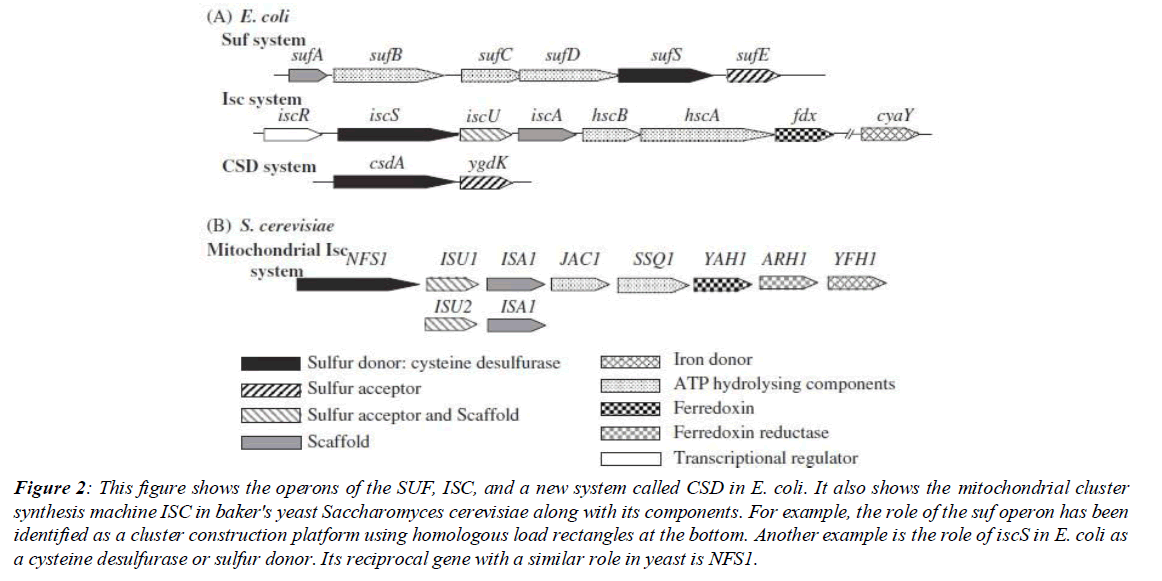
Molecular genetic research on iron-sulfur clusters started with work on nitrogenase in Azotobacter vinelandii. Mutations in the nifSU genes inhibited nitrogenase maturation. Nitrogenase is a protein enzyme with a high number of subunits that plays a role in nitrogen fixation in plants in low oxygen conditions. These mutations provided incentives. These incentives paved the way to investigate the genes that had access to the nitrogenase cofactors. The interesting thing was that disabling the nifSU gene did not eliminate all nitrogenase activity. It was clear that other genes were involved in nitrogenase activity. Interestingly, after these studies, which started in the late 1980s, the isc genes were discovered (Figure 2).
Figure 2: This figure shows the operons of the SUF, ISC, and a new system called CSD in E. coli. It also shows the mitochondrial cluster synthesis machine ISC in baker's yeast Saccharomyces cerevisiae along with its components. For example, the role of the suf operon has been identified as a cluster construction platform using homologous load rectangles at the bottom. Another example is the role of iscS in E. coli as a cysteine desulfurase or sulfur donor. Its reciprocal gene with a similar role in yeast is NFS1.
Research was mainly focused on two organisms - Escherichia coli and baker's yeast. However, phylogenetic analysis by (Tokumoto et al., 2004) showed that in most classes of proteobacteria (gamma and beta), the cluster synthesis system exists. Here we refer to Mr. Tokumoto's article. Early research indicated the existence of three main systems: (Iron-sulfur Cluster) ISC, (Sulfur mobilization) SUF and NIF (Nitrogen fixation). The NIF system has two gene groups (nifU) and (nifS). The first group, as a scaffold protein, provides a platform for the synthesis of iron-sulfur clusters from its precursors. The second group is a cysteine desulfurase (removal of sulfur from the amino acid cysteine and its conversion to alanine) that is dependent on pyridoxal phosphate (a cofactor) and initiates cluster formation by removing sulfur from cysteine and transferring it to the first group. This information was published in a scientific report by the Dean group. In bacteria, genes with similar (not always) functions are placed in units called operons. The isc genes in bacteria are located in the (iscSUA-hscBA-fdx) operon. This operon contains eight genes. Mutation in this operon causes decreased activity of many cluster proteins. Next, their functions and reciprocal genes in eukaryotes will be discussed. Subsequently, a new system (SUF) was discovered in bacteria. This system is encoded by the (sufABCDSE) operon and is considered an alternative pathway for cluster synthesis. Another question here was how was the role of the SUF system in cluster synthesis confirmed? The answer was that in a mutated E. coli, the entire isc operon was disabled. However, the activity of iron-sulfur proteins was determined to be about 2 to 10 percent of normal. The remaining activity was derived from other systems. To examine the role of the new system, expression of the suf operon was increased and it was observed that the activity of iron-sulfur proteins was partially recovered.
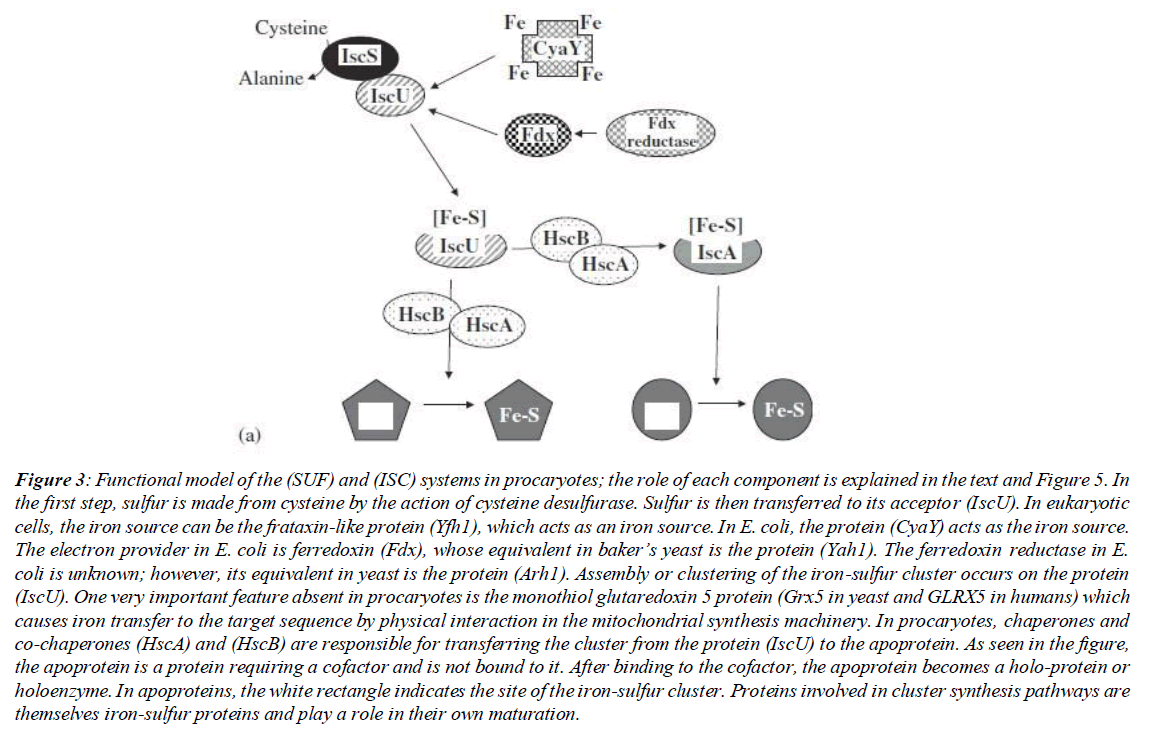
The iscS gene is responsible for taking sulfur from cysteine and donating it to the sulfur acceptor. As a result of deleting this gene from the E. coli isc operon, the cells show less growth compared to the wild strain. Also, as a result of this deletion mutation, the cell in the aerobic environment and with the minimum requirements for growth, requires thiamine (vitamin B1) and nicotinic acid (niacin; a form of vitamin B3). In anaerobic environments, cells only require niacin. The thiamine intake deficiency may be due to a defect in iron-sulfur cluster synthesis, while the requirement for niacin may indicate a deficiency in NAD biosynthesis. Niacin is not a direct precursor of NAD. However, niacin can bypass its synthesis pathway immediately after the action of the NadA quinolinate synthase enzyme. This enzyme contains an iron-sulfur cluster. Other studies have shown that a defect in the related gene causes decreased activity in several important enzymes such as succinate dehydrogenase, NADH dehydrogenase and glutamate synthase. All these sources discuss iscS as a major source of sulfur for iron-sulfur cluster biosynthesis. After sulfur separation by the cysteine desulfurase system from cysteine, the next step is the receipt of sulfurs by a sulfur acceptor [2]. This action in different E. coli systems is performed by two genes, sufE and iscU. An interesting point about these sulfur acceptors is the structural similarities between the two sulfur acceptors. (Figure 3)
Figure 3: Functional model of the (SUF) and (ISC) systems in procaryotes; the role of each component is explained in the text and Figure 5. In the first step, sulfur is made from cysteine by the action of cysteine desulfurase. Sulfur is then transferred to its acceptor (IscU). In eukaryotic cells, the iron source can be the frataxin-like protein (Yfh1), which acts as an iron source. In E. coli, the protein (CyaY) acts as the iron source. The electron provider in E. coli is ferredoxin (Fdx), whose equivalent in baker’s yeast is the protein (Yah1). The ferredoxin reductase in E. coli is unknown; however, its equivalent in yeast is the protein (Arh1). Assembly or clustering of the iron-sulfur cluster occurs on the protein (IscU). One very important feature absent in procaryotes is the monothiol glutaredoxin 5 protein (Grx5 in yeast and GLRX5 in humans) which causes iron transfer to the target sequence by physical interaction in the mitochondrial synthesis machinery. In procaryotes, chaperones and co-chaperones (HscA) and (HscB) are responsible for transferring the cluster from the protein (IscU) to the apoprotein. As seen in the figure, the apoprotein is a protein requiring a cofactor and is not bound to it. After binding to the cofactor, the apoprotein becomes a holo-protein or holoenzyme. In apoproteins, the white rectangle indicates the site of the iron-sulfur cluster. Proteins involved in cluster synthesis pathways are themselves iron-sulfur proteins and play a role in their own maturation.
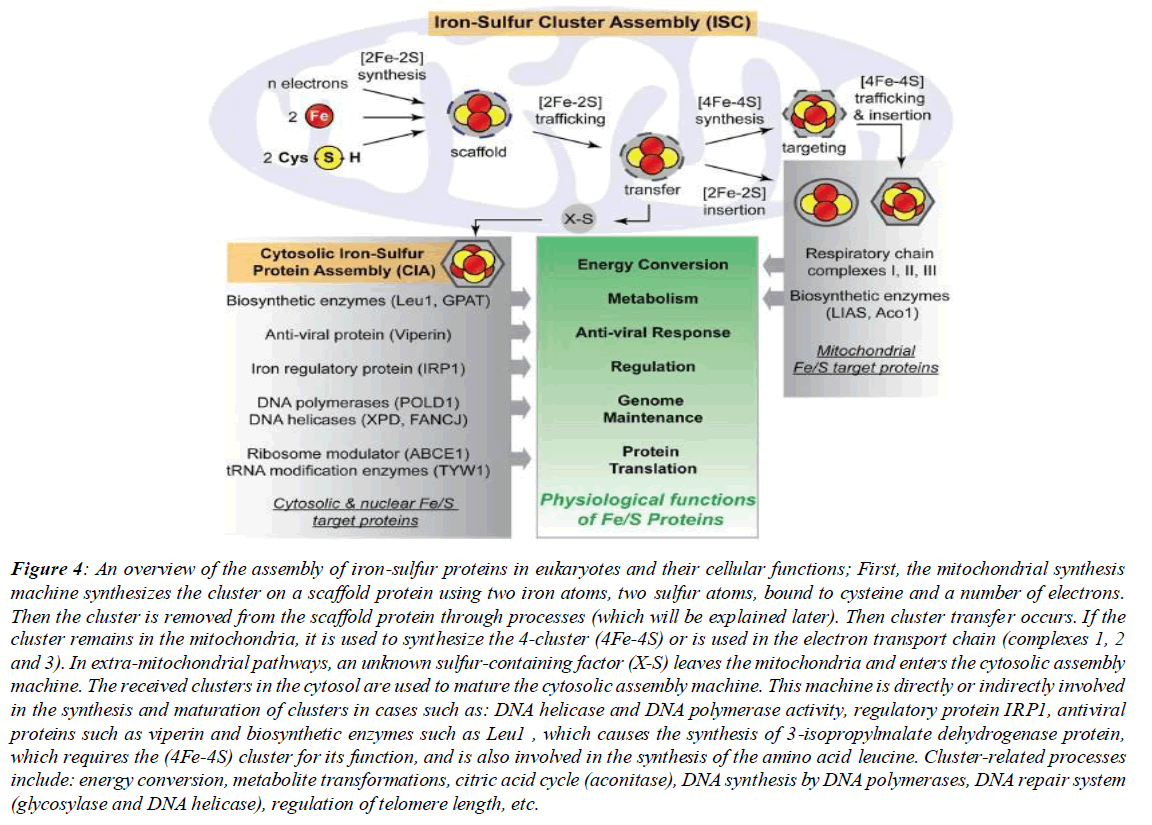
The role of the SUF system is more pronounced in stresses such as oxidative stresses. Interestingly, analysis and examination of the expression level of the suf operon gene has shown that this operon is controlled by a regulator sensitive to hydrogen peroxide. With this in mind, it should be said that the role of this system is more pronounced in iron limitations and oxidative stresses. For example, a mutation in sufC causes decreased activity of several iron-sulfur proteins such as 6-phosphogluconate dehydrogenase and fumarase A during oxidative stress. It is clearly evident that sufC plays a regulatory role under specific conditions for these enzymes. In (Tokumoto et al., 2004), the complete or partial deletion of operons by plasmid and other tools and the role of other operon genes are discussed [3]. Those interested can refer to the aforementioned reference at the end of the text for more information. (Figure 4)
Figure 4: An overview of the assembly of iron-sulfur proteins in eukaryotes and their cellular functions; First, the mitochondrial synthesis machine synthesizes the cluster on a scaffold protein using two iron atoms, two sulfur atoms, bound to cysteine and a number of electrons. Then the cluster is removed from the scaffold protein through processes (which will be explained later). Then cluster transfer occurs. If the cluster remains in the mitochondria, it is used to synthesize the 4-cluster (4Fe-4S) or is used in the electron transport chain (complexes 1, 2 and 3). In extra-mitochondrial pathways, an unknown sulfur-containing factor (X-S) leaves the mitochondria and enters the cytosolic assembly machine. The received clusters in the cytosol are used to mature the cytosolic assembly machine. This machine is directly or indirectly involved in the synthesis and maturation of clusters in cases such as: DNA helicase and DNA polymerase activity, regulatory protein IRP1, antiviral proteins such as viperin and biosynthetic enzymes such as Leu1 , which causes the synthesis of 3-isopropylmalate dehydrogenase protein, which requires the (4Fe-4S) cluster for its function, and is also involved in the synthesis of the amino acid leucine. Cluster-related processes include: energy conversion, metabolite transformations, citric acid cycle (aconitase), DNA synthesis by DNA polymerases, DNA repair system (glycosylase and DNA helicase), regulation of telomere length, etc.
In Eukaryotes
Research on the assembly and construction of clusters in eukaryotes has been mainly conducted on the model organism, Saccharomyces cerevisiae. The functioning of many cellular proteins depends on the cofactors that form part of their active site. In this case, different cells or species carry out various strategies to import or synthesize these cofactors. Inorganic iron-sulfur clusters are highly dependent on the intracellular environment for their synthesis. This fact indicates that unbound iron-sulfur clusters are unstable in aqueous solutions. This stability is generally achieved through coordination with histidine and cysteine ligand arms. Most species of all animal kingdoms contain iron-sulfur clusters and are often dependent on them for survival. Exceptions are seen in some strains of Mycoplasma and Borrelia burgdorferi (Figure 5).
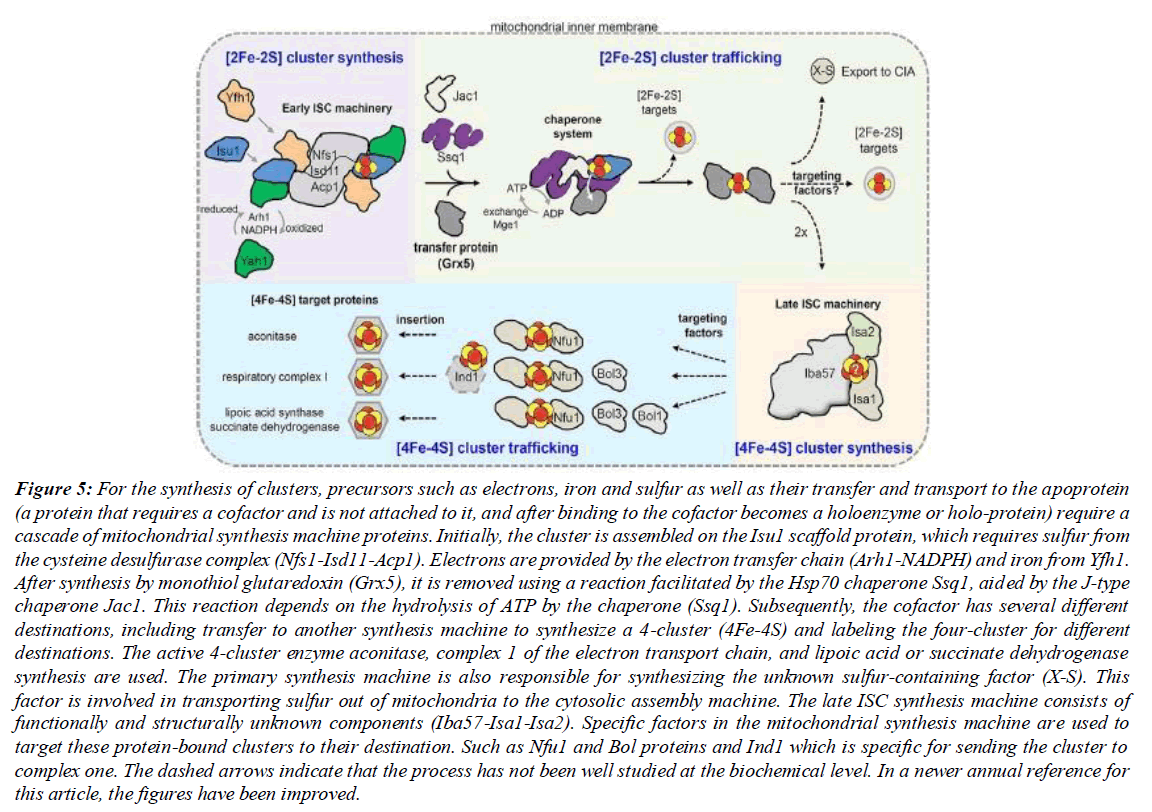
Figure 5: For the synthesis of clusters, precursors such as electrons, iron and sulfur as well as their transfer and transport to the apoprotein (a protein that requires a cofactor and is not attached to it, and after binding to the cofactor becomes a holoenzyme or holo-protein) require a cascade of mitochondrial synthesis machine proteins. Initially, the cluster is assembled on the Isu1 scaffold protein, which requires sulfur from the cysteine desulfurase complex (Nfs1-Isd11-Acp1). Electrons are provided by the electron transfer chain (Arh1-NADPH) and iron from Yfh1. After synthesis by monothiol glutaredoxin (Grx5), it is removed using a reaction facilitated by the Hsp70 chaperone Ssq1, aided by the J-type chaperone Jac1. This reaction depends on the hydrolysis of ATP by the chaperone (Ssq1). Subsequently, the cofactor has several different destinations, including transfer to another synthesis machine to synthesize a 4-cluster (4Fe-4S) and labeling the four-cluster for different destinations. The active 4-cluster enzyme aconitase, complex 1 of the electron transport chain, and lipoic acid or succinate dehydrogenase synthesis are used. The primary synthesis machine is also responsible for synthesizing the unknown sulfur-containing factor (X-S). This factor is involved in transporting sulfur out of mitochondria to the cytosolic assembly machine. The late ISC synthesis machine consists of functionally and structurally unknown components (Iba57-Isa1-Isa2). Specific factors in the mitochondrial synthesis machine are used to target these protein-bound clusters to their destination. Such as Nfu1 and Bol proteins and Ind1 which is specific for sending the cluster to complex one. The dashed arrows indicate that the process has not been well studied at the biochemical level. In a newer annual reference for this article, the figures have been improved.
Examples of the most abundant iron-sulfur clusters in living cells include [4Fe-4S], [3Fe-4S], [2Fe-2S]. While these compounds have abbreviated and simple nomenclature, the synthesis, packaging, assembly of these proteins, and ultimately their delivery to apoproteins is facilitated by highly complex protein machines. The fact that the synthesis of iron-sulfur clusters is a process catalyzed by enzymes was discovered in 1999. Shortly before that, the identification of the bacterial operon (isc) took place. The synthesis of proteins by this bacterial operon was related to the process of synthesizing iron-sulfur clusters in mitochondria. Initial studies were conducted on baker's yeast, Saccharomyces cerevisiae. However, later research expanded in different branches of eukaryotes, and subsequent findings indicated a very strong preservation of the iron-sulfur cluster synthesis pathway from yeast to humans. A combination of studies in the laboratory (artificial) and natural environments currently indicates a set of specific events that are interconnected for the synthesis of iron-sulfur clusters in the cytosol, mitochondria and nucleus. In summary, the assembly of [4Fe-4S], [2Fe- 2S] clusters and their attachment to cellular apoproteins is catalyzed by more than 30 proteins.
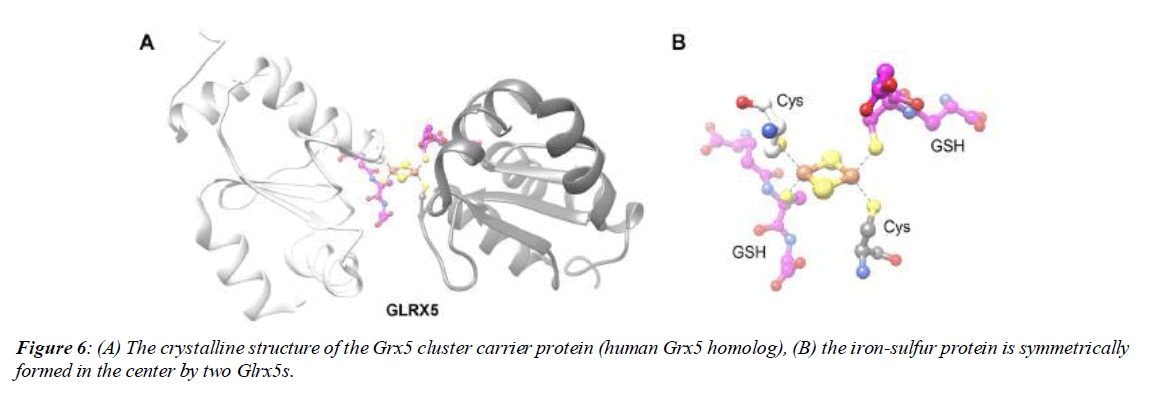
The cluster synthesis process in the mitochondrial matrix is initiated by 18 proteins in the form of a mitochondrial cluster synthesis system. This machine is thought to have been inherited from bacteria to eukaryotes through endosymbiosis. Cluster synthesis occurs in three locations - mitochondria, cytosol and nucleus - to employ clusters in different roles. The mitochondrial synthesis machine is not only responsible for synthesizing its own organelle proteins, but also plays an important role in synthesizing all extra-mitochondrial Fe-S proteins. Of course, there are exceptions to this, such as epicoplastic proteins that mature through the SUF chloroplastic system (Figure 6A & 6B).
The central part of the mitochondrial synthesis machine must subsequently produce a sulfur-containing factor, which is transported to the cytosol by the ABCB7 transporter. Afterwards, the sulfur factor is used by the Cytosolic Iron- Sulfur Protein Assembly machinery, briefly called CIA. This machine consists of 11 known proteins for the construction of nuclear and cytosolic iron-sulfur proteins.
The specific Hsp70 chaperone, Ssq1, and its co-chaperone Jac1 together target the Isu1 scaffold protein. This targeting is to release the assembled iron-sulfur cluster from the scaffold protein and transfer it to monothiol glutaredoxin (Grx5). Subsequently, Jac1 detects the Isu1-bound cluster and directs it toward Ssq1, also facilitating the separation of the cluster from Isu1. Grx5 is the only known protein capable of directly binding to the iron-sulfur cluster through physical interaction with Ssq1. In summary, Jac1 plays a role in guiding the Isu1-cluster complex toward Ssq1. Also, Ssq1 plays the role of interacting with the Isu1-cluster complex and Grx5. And Grx5 receives the cluster by physical reaction with the nonsubstrate active site of Ssq1 and plays a role in cluster transfer or trafficking. These findings were obtained through research and analysis from artificial environments utilizing radioactive labeling of clusters of the mitochondrial synthesis machinery. In general, the chaperone system is comprised of the components Ssq1, Jac1, and Grx5. Collectively they constitute the chaperone complex. Within this complex, Grx5 functions as a cluster [2Fe-2S] transfer protein. Symmetrically, two monomers are attached to a [2Fe-2S] cluster and play a role in transporting it. The principal component of the mitochondrial synthesis machinery is cysteine desulfurase or NFS1, which by converting cysteine to alanine, provides the sulfur requisite for cluster construction. Dimerization (existing as two-fold) of the protein induces this complex to assume a symmetrical form.
This structure was attained utilizing cryo-electron microscopy and X-ray crystallography, nuclear magnetic resonance. In the spatial position above NFS1, ISD11 proteins are connected to each other and have an interaction or reciprocal reaction. Each of the ISD11s in a duplicative manner on the previous states can attach to the acyl carrier protein or ACP1. The ACP1 protein is one of the key proteins in the second system of mitochondrial fatty acid synthesis [4]. ATP hydrolysis on ssq1 induces a conformational change in the second attachment to the ssq1 peptide, which leads to its guidance toward stable attachment to the LPPV motif from Isu1. It is also believed that this reaction marks the existing cluster on Isu1 so that like a relay its attachment to GRX5 is facilitated. When the cluster release process has occurred once, the nucleotide exchange factor or Mge1 can restore Ssq1 (the ability to rebind ATP). The regulated steps of the chaperone cycle can prevent the unintentional release of iron-sulfur clusters into solutions. In support of this perspective, deletion of the chaperone system or Grx5 in the intracellular organism environment shows an increase in iron-sulfur clusters bound to Isu1. (Figure 7)
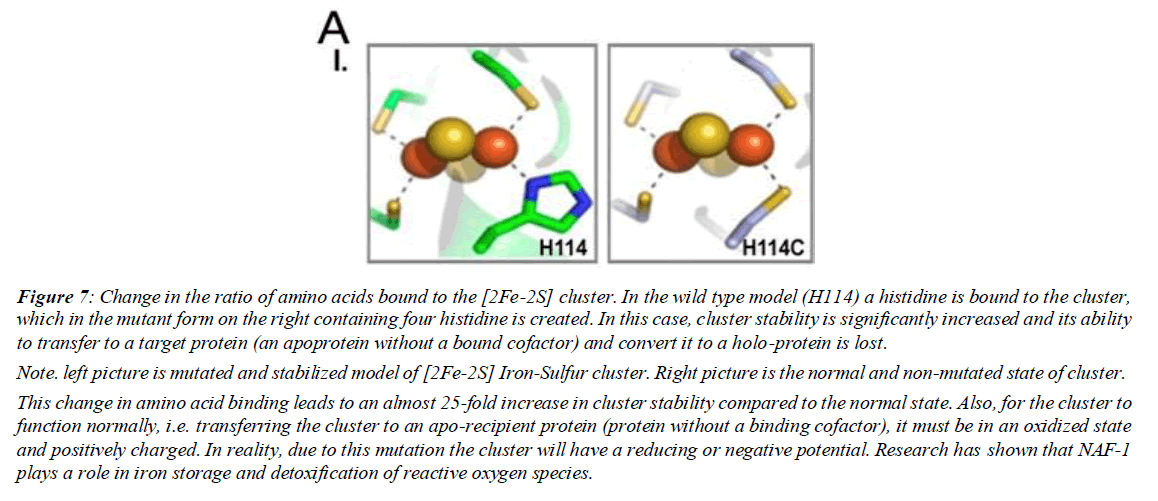
Figure 7: Change in the ratio of amino acids bound to the [2Fe-2S] cluster. In the wild type model (H114) a histidine is bound to the cluster, which in the mutant form on the right containing four histidine is created. In this case, cluster stability is significantly increased and its ability to transfer to a target protein (an apoprotein without a bound cofactor) and convert it to a holo-protein is lost.
Note. left picture is mutated and stabilized model of [2Fe-2S] Iron-Sulfur cluster. Right picture is the normal and non-mutated state of cluster.
This change in amino acid binding leads to an almost 25-fold increase in cluster stability compared to the normal state. Also, for the cluster to function normally, i.e. transferring the cluster to an apo-recipient protein (protein without a binding cofactor), it must be in an oxidized state and positively charged. In reality, due to this mutation the cluster will have a reducing or negative potential. Research has shown that NAF-1 plays a role in iron storage and detoxification of reactive oxygen species.
The relationship between cluster stability and deficiency with cancer
NAF-1 is a new group from the NEET family of ironsulfur proteins. This protein family is encoded by the cisd gene family. The role of this group in autophagy and other processes such as programmed cell death and altering cellular resistance to oxidative stress. Also, the effect of this iron-sulfur cluster carrying factor on the tumor volume of breast cancer in cell culture using the MDA-MB-231 cell line has been investigated. During experiments using the aforementioned cell line, it was examined whether there is a relationship between NAF-1 and tumor growth rate. NAF-1 expression was examined in two states: overexpression and suppressing protein function by increasing iron-sulfur cluster stability. The first state leads to increased detoxification of reactive oxygen species, increased pentose phosphate pathway flux in the form of increased NADPH reserves, decreased oxidation of fatty acids, and other effects. Ultimately the first state leads to increased cellular resistance to oxidative stress. The second state, or increasing iron-sulfur cluster stability, occurs via introducing a point mutation in the sequence encoding the iron-sulfur protein. This mutation changes the coordination structure of the iron-sulfur cluster from 3-cysteine 1-histidine to 4-cysteine.
This change in amino acid binding leads to an almost 25-fold increase in cluster stability compared to the normal state. Also, for the cluster to function normally, i.e. transferring the cluster to an apo-recipient protein (protein without a binding cofactor), it must be in an oxidized state and positively charged. In reality, due to this mutation the cluster will have a reducing or negative potential. Research has shown that NAF- 1 plays a role in iron storage and detoxification of reactive oxygen species [5].
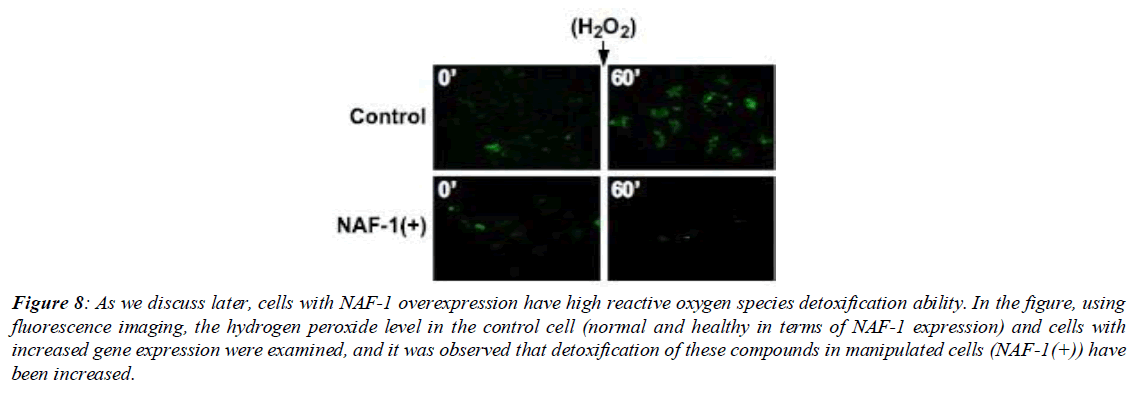
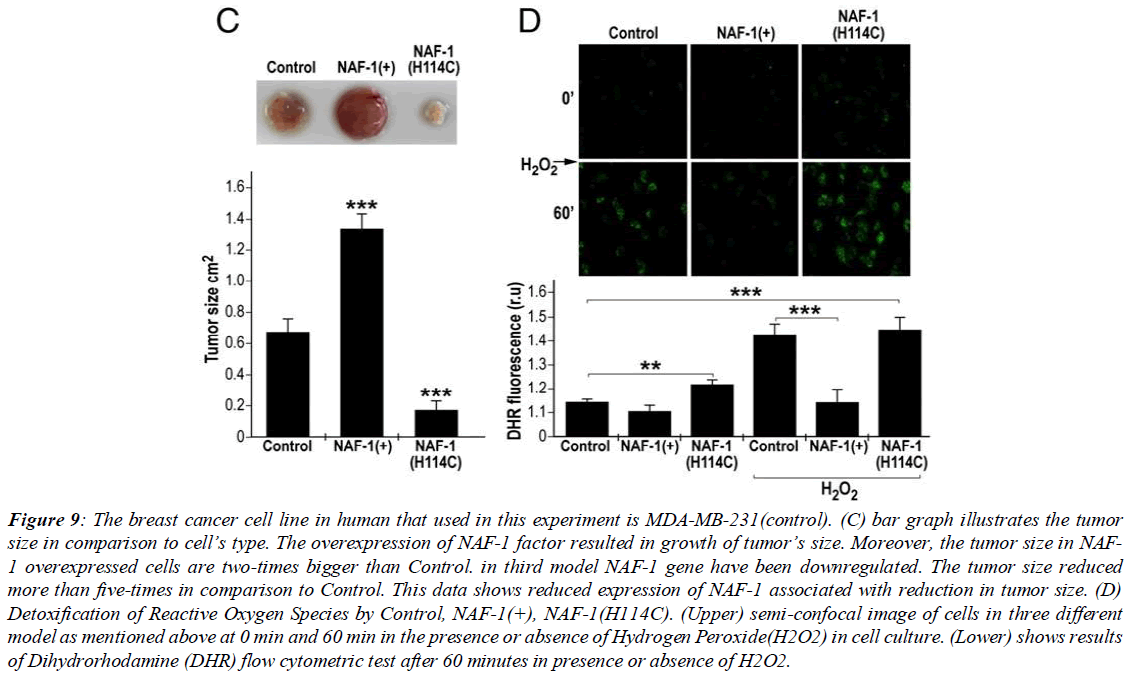
In examining the role of iron-sulfur clusters in oxidative stress and cancer tumor growth, Darash-Yahana et al. defined three models: NAF-1(+), NAF-1(H114C), and Control, which respectively in the first model protein expression is increased. in the second model a point mutation has occurred that causes cluster stability and deficiency. the third model (Control) remained without change. Darash-Yahana et al. transferred each of these models to a separate environment and added hydrogen peroxide (H2O2) to the environment of all three. In the next step, the performance of the three models in combating oxidative radical compounds after exposure to hydrogen peroxide was examined using confocal microscopy. The examinations showed that cells containing the mutant protein NAF-1(H114C) did not have the ability to detoxify hydrogen peroxide and reserves of reactive oxygen species in them increased markedly. In contrast, cells containing the overexpressed protein NAF-1(+) had the ability to detoxify the aforementioned compound (Figure 8).
Figure 8: As we discuss later, cells with NAF-1 overexpression have high reactive oxygen species detoxification ability. In the figure, using fluorescence imaging, the hydrogen peroxide level in the control cell (normal and healthy in terms of NAF-1 expression) and cells with increased gene expression were examined, and it was observed that detoxification of these compounds in manipulated cells (NAF-1(+)) have been increased.
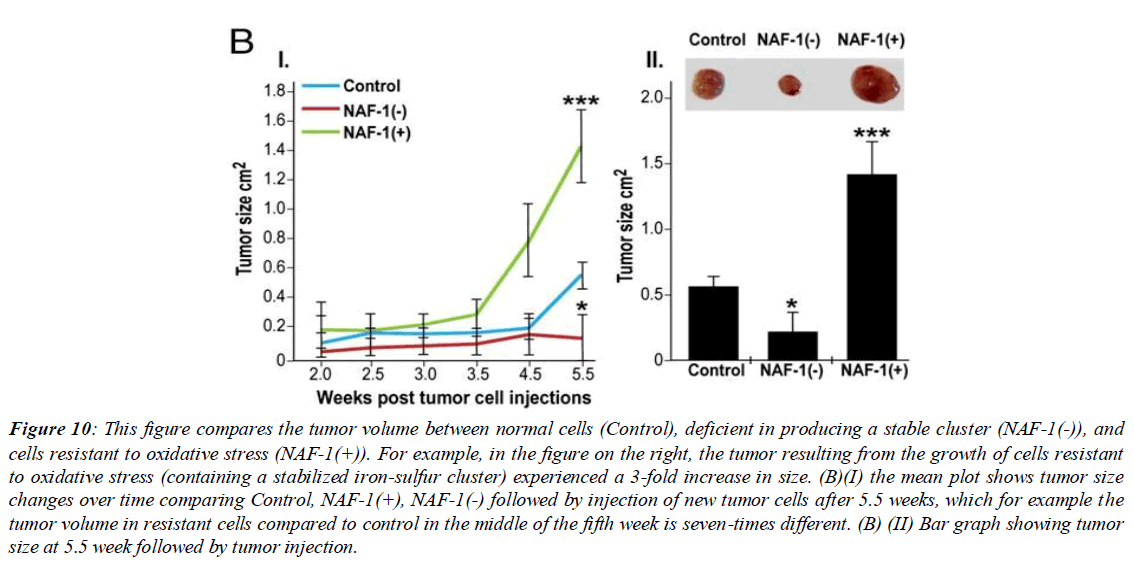
The relationship between cluster stability and tumor growth in the aforementioned article was examined, and it was observed that the increased stability in the cluster led to a deficiency in cluster protein transfer, and subsequently, due to lack of cluster transfer, cell resistance against oxidative stress decreases, and tumor growth volume markedly (more than one third compared to control) decreases. Pioglitazone has a similar effect as the NAF-1 point mutation in cancer cells, in that it causes stability and suppresses NAF-1 function in cancer cells (Figure 9). The thiazolidinedione drug class PPARγ (peroxisome proliferator-activated receptor gamma) includes antidiabetic drugs that reduce insulin resistance. Pioglitazone, one type of thiazolidinedione, inhibits the ability of the NAF-1 cluster protein to transfer its cluster to apo-recipient proteins. In previous research it has been observed that pioglitazone decreases the rate of cell division by inducing cell death across a wide spectrum of cancer cell types. Also, in examining the effect of pioglitazone on mRNA expression levels of gamma and alpha peroxisome proliferator-activated receptors, studies were conducted on MDA-MB-231 cells and examining the effect of the drug on them. The results obtained showed that treating the desired cell line with 30 micromoles pioglitazone, increased PPAR gamma receptor mRNA expression by about 4-fold compared to control after 6 days [6]. Additionally, in another part of the research it was observed that the ability of cells undergoing pioglitazone treatment to survive decreased to a minimum level, and the results showed no correlation between PPAR gamma receptor mRNA expression levels and other isoforms with the ability of cells undergoing PGZ treatment to survive. In determining whether apoptosis occurred, using realtime PCR technique, mRNA expression of caspases 9 and 3 were examined, and it was observed that the caspases were undetectable. This observation was confirmation that apoptosis of cancer cells using pioglitazone occurs through other cellular mechanisms that are undetectable by DNA ladder technique. (Figure 10)
Figure 9: The breast cancer cell line in human that used in this experiment is MDA-MB-231(control). (C) bar graph illustrates the tumor size in comparison to cell’s type. The overexpression of NAF-1 factor resulted in growth of tumor’s size. Moreover, the tumor size in NAF- 1 overexpressed cells are two-times bigger than Control. in third model NAF-1 gene have been downregulated. The tumor size reduced more than five-times in comparison to Control. This data shows reduced expression of NAF-1 associated with reduction in tumor size. (D) Detoxification of Reactive Oxygen Species by Control, NAF-1(+), NAF-1(H114C). (Upper) semi-confocal image of cells in three different model as mentioned above at 0 min and 60 min in the presence or absence of Hydrogen Peroxide(H2O2) in cell culture. (Lower) shows results of Dihydrorhodamine (DHR) flow cytometric test after 60 minutes in presence or absence of H2O2.
Figure 10: This figure compares the tumor volume between normal cells (Control), deficient in producing a stable cluster (NAF-1(-)), and cells resistant to oxidative stress (NAF-1(+)). For example, in the figure on the right, the tumor resulting from the growth of cells resistant to oxidative stress (containing a stabilized iron-sulfur cluster) experienced a 3-fold increase in size. (B)(І) the mean plot shows tumor size changes over time comparing Control, NAF-1(+), NAF-1(-) followed by injection of new tumor cells after 5.5 weeks, which for example the tumor volume in resistant cells compared to control in the middle of the fifth week is seven-times different. (B) (ІІ) Bar graph showing tumor size at 5.5 week followed by tumor injection.
Conclusion
In examining two related cancer articles, it was determined that stabilizing the cluster in two ways - pioglitazone and point mutation - reduces cell resistance to oxidative stress. This is because the iron-sulfur cluster in a normal cell may become defective or its stability may be artificially reduced by drugs. This stability reduction leads to reduced cell resistance against oxidative stress. One characteristic of cancer cells is their high metabolism. This high metabolism leads to high levels of reactive oxygen species production. However, cancer cells have adapted to detoxify these active compounds. One new cancer treatment is the use of iron nano-catalysts, where labeled iron enters the tumor tissue and causes high levels of reactive oxygen species production. Cancer cells are incompetent in fighting these active species, and this method can be effective in treating cancer. The relationship between iron-sulfur clusters and cancer treatment is simple. If the cancer cell is resistant to oxidative stress and has the unusual ability to detoxify active oxygen compounds, cluster destabilization can be performed. As an example, as in breast cancer, by creating a mutation in the NAF-1 gene, the stabilized cluster causes the cell to lose its ability to fight reactive oxygen compounds, and tumor volume markedly decreases. Also, by adding hydrogen peroxide, no noticeable decrease is observed in its level in these mutant cells. Here, it can be concluded that making changes in iron-sulfur clusters, whether directly by injecting hydrogen peroxide into the culture medium or indirectly through Fenton reactions and iron nano-catalysts, can exploit these methods to treat cancer.
References
- Beinert H, Holm RH, Munck E. Iron-sulfur clusters: Nature's modular, multipurpose structures. Science. 1997;277(5326):653-9.
- Barras F, Loiseau L, Py B. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv Microb Physiol. 2005;50:41-101.
- Tokumoto U, Kitamura S, Fukuyama K, et al. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J Biochem. 2004;136(2):199-209.
- Braymer JJ, Lill R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017;292(31):12754-63.
- Darash-Yahana M, Pozniak Y, Lu M, Sohn YS, et al. Breast cancer tumorigenicity is dependent on high expression levels of NAF-1 and the lability of its Fe-S clusters. Proc Natl Acad Sci. 2016;113(39):10890-5.
- Nadarajan K, Balaram P, Khoo BY. MK886 inhibits the pioglitazone-induced anti-invasion of MDA-MB-231 cells is associated with PPARα/γ, FGF4 and 5LOX. Cytotechnology. 2016;68:1771-87.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref