Research Article - Biomedical Research (2018) Volume 29, Issue 4
Identification of SIRT4 interacting proteins in mouse brain
Gizem Yalcin* and Abdullah Yalcin
Department of Medical Biology, Faculty of Medicine, Adnan Menderes University, Aydin, Turkey
- *Corresponding Author:
- Gizem Yalcin
Department of Medical Biology
Faculty of Medicine
Adnan Menderes University
Turkey
Accepted date: December 04, 2017
DOI: 10.4066/biomedicalresearch.29-17-3585
Visit for more related articles at Biomedical ResearchAbstract
Sirtuins are enzymes that respond to cellular or organismal stress by activation or inhibition of the activities of proteins via deacetylating or ADP-ribosylating them after translation. Previously, it was shown that sirtuins affect the age-related pathways in the cell via interfering with the metabolism. SIRT4 was also shown to affect the glutamate metabolism both in pancreatic cells and also in brain. The aim of this study is to identify the interacting proteins of mitochondrial SIRT4 in mouse brain. We identified a number of proteins that interact with SIRT4 in mouse brain. We also showed that the ATP levels are lower in SIRT4 KO mouse brain compared to controls. Revealing SIRT4 interacting proteins will help to understand its function better in brain and lead the path for developing therapies for agerelated brain diseases.
Keywords
Protein, Mass spectrometry, Chaperone, Mitochondria, SIRT4
Introduction
Sirtuins are expressed in different cellular compartments including nucleus, cytoplasm and mitochondria [1]. They post-translationally modify proteins via deacetylation, ADP-ribosylation, malonilation, succinylation [2]. These functions of sirtuins lead to the activation or inhibition of different proteins and enzymes. Sirtuins were shown to play roles in many metabolic pathways related to aging such as brain diseases [1,3]. Sirtuins are activated as a result of cellular or organismal stress or injury.
There are seven sirtuin homologs known as SIRT1-7. Among those, SIRT3-5 are expressed in mitochondria [2]. The function and substrates of mitochondrial SIRT4 are not clearly known. It is known that SIRT4 is a weak deacetylase and also an ADP-ribosylase enzyme [3]. The only known substrate for SIRT4 was found to be Glutamate Dehydrogenase (GDH) in pancreatic cells [3]. With this study, the relation between SIRT4 and glutamate metabolism was uncovered and it was thought that there is a high probability that it has a similar role in brain. Another study had found that SIRT4 was expressed in the CTX8 radial glial cell line and SIRT4 and glutamate dehydrogenase 1 regulate glial development [4]. For this reason, it will also be interesting to study the function of SIRT4 in brain and its relation to brain diseases.
In a recent study, the relation between SIRT4 and Glutamate Transporter 1 (GLT-1) was uncovered in mouse brain [5]. Excitotoxicity forms as a result of accumulation of excessive glutamate in synapse between two neurons. Excess glutamate is mostly absorbed by presynaptic astrocyte. This clearance mechanism is extremely important [6]. Mostly, receptors called GLT-1 take up the excess glutamate from synapses. Excitotoxicity affects the mitochondria function, since excessive glutamate disrupts Ca+2 balance and ATP production and further leads to the formation of reactive oxygen species. It is associated with age-related neurodegenerative diseases and also stroke, epilepsy and traumatic brain injury [7,8]. Preventing excitotoxicity will help to stop or slow the progress of these diseases. Kainic acid is an AMPA/kainate receptor agonist in brain and a potent excitotoxin [5]. It induces seizure phenotype, glutamate increase in synapse, and cell death. Shih et al. showed that SIRT4 KO mice have enhanced seizure phenotypes compared to wild type mice after kainic acid injection, and that in wild-type mice, SIRT4 levels were increased in the hippocampus 24 h after treatment. It was also demonstrated that loss of SIRT4 decreases GLT-1-dependent glutamate uptake and increases sensitivity to kainic acid and induces stress response [5].
In the light of the previous findings, it is clear that SIRT4 has important functions in brain. In this study, we first showed that SIRT4 affects ATP production in mouse brain, since ATP levels were decreased in SIRT4 KO mouse brain extract compared to wild type mouse. We then identified the SIRT4 interacting proteins in mouse brain via immunoprecipitation and mass spectrometry. Target proteins identified are HSC71, calreticulin, GRP78 (78 kDa glucose regulated protein), annexin A5 and peroxiredoxin 1. These proteins play roles as chaperones, or in Ca+2 balance and redox regulation. They are all associated with mitochondria functions and stress response. It will be interesting in the future to detail the interactions of SIRT4 with these proteins and related metabolic pathways. This will broaden our understanding about the role of SIRT4 in brain glutamate metabolism, excitotoxicity, brain function and diseases. It will also help develop therapeutic strategies for brain diseases.
Materials and Methods
Mice
Wild-type (129S3/SvImJ) and SIRT4 KO (129-Sirt4<tm1Fwa>/J) mice were obtained from Jackson Laboratories (Bar Harbor, ME, USA). All animal procedures were performed in accordance with the guidelines and approved by Institutional Animal Care and the relevant Institutional Ethical Review Committees.
Preparation of the mouse brain extracts
Wild type and SIRT4 knockout mice were anesthetized and sacrificed by cervical dislocation, and their brains were removed immediately. Half brains were digested in RIPA lysis and extraction buffer according to the manufacturer’s guidelines (Thermo Fisher Scientific).
ATP measurement
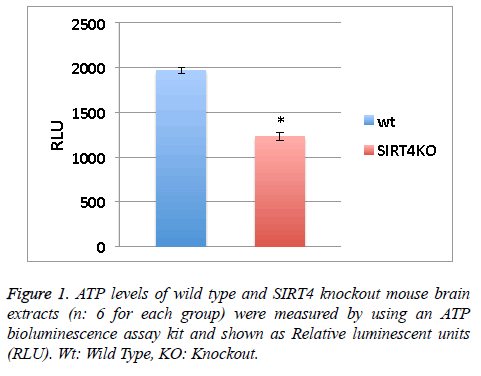
ATP levels of wild type and SIRT4 knockout mouse brain extracts (n: 6 for each group) were measured by using an ATP bioluminescence assay kit according to the manufacturer’s guidelines (Roche Diagnostics). Relative Luminescent Units (RLU) from 6 measurements were averaged (Figure 1).
Isolation of proteins
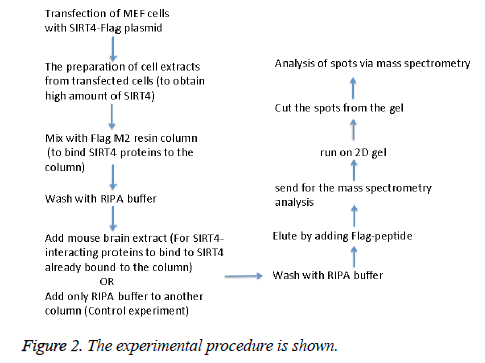
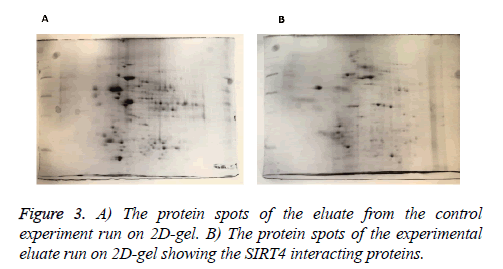
MEFs (mouse embryonic fibroblasts) were transfected with SIRT4-FLAG plasmid (Addgene) using Lipofectamine according to the manufacturer’s protocol (Thermo Fisher Scientific). Figure 2 shows the experimental procedure. Total cell extracts from SIRT4 transfected MEFs were prepared using RIPA lysis and extraction buffer (Thermo Fisher Scientific) and 1 ml of the extract was run through 20 μl of anti-FLAG M2 resin (Sigma-Aldrich) column. As a control experiment, RIPA buffer was run through a second column of anti-FLAG M2 resin instead of cell extract (Figure 3A). After washing with PBS, mouse brain extracts (1 ml of 1 mg/ml) were passed through both of the columns. Then, the proteins bound to the both columns were eluted by passing FLAG peptide through both of the columns. Both eluates (experiment and the control) were frozen and sent to Kocaeli University, Faculty of Medicine, DEKART Proteomics Laboratory for Mass Spectrometry analysis.
Mass spectrometry
a) Sample preparation: The frozen eluates were thawed on ice and the protein concentrations were measured using Bradford assay (BioRad, USA).
b) 2D- gel electrophoresis (2-DE): Immobilized pH gradient (IPG) strips (17 cm, pH 3-10, BioRad, USA) were passively rehydrated using 2D-Rehydration buffer, which contains 500 µg of protein at 20°C for 16 h. Then, the strips were run through a stepwise incremental voltage program (250 V for 20 min (linear), 4000 V for 2 h (linear) and 40,000 V/h (rapid)) using a Protean IEF system (BioRad, USA) and subjected to a two-step equilibration in buffers containing 6 M urea, 2% SDS, 0.375 M Tris. Cl at pH 8.8, 20% glycerol and 2% DTT for the first step. For the second step, the same buffer was used without DTT but with iodoacetamide (2.5%). The strips were then subjected to second dimension separation using SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE) (12%). 40% Methanol and 10% acetic acid were used to fix the gels. Then, gels were stained with colloidal coomassie stain (KeraFast, USA), and visualized with VersaDoc4000MP (Bio-Rad, USA).
c) Image analysis and statistical significance: PD Quest Advanced Software (BioRad, USA) was used to analyse the images and the automated crop tool was used to identically crop the outer edges of the images. Stain speckles were filtered and the standardized areas of interest from all gels were matched and warped. The total valid spot intensity was used to normalize the quantity of each spot. Total spot numbers and volumes within the normalized area were determined from the automated analyses. The determined protein spots detected by the software were inspected using a manual editing tool. Spots that were prone to variation were excluded if the spots were hard to identify by visual inspection. Every-member matching protein spots were selected. An automated spot cutting tool, ExQuest spot cutter (Bio-Rad, USA), was used to cut the spots. The spots were then disposed into a 96-well plate for protein identification.
d) In-gel tryptic digestion: An in-gel digestion kit was used to perform the in-gel tryptic digestions of the proteins according to the protocol by the manufacturer (Pierce, USA). The selected protein spots were de-stained/washed with 40% acetonitrile and 50 mM ammonium bicarbonate until the gel pieces lost their color. They were then reduced with 10 mM DTT at 60°C for 30 min and alkylated in the dark with 50 mM iodoacetamide at room temperature for 30 min. 200 μl acetonitrite was used to dehydrate the gel pieces for 15 min with shaking. Acetonitrite was removed and the gel pieces were dried at room temperature after dehydration. 10 ng trypsin in 20 μl 40 mM NH4HCO3 solution was added onto each spot followed by incubation at 37°C for overnight for tryptic digestion. The peptides were then collected, evaporated in a SpeedVac (Eppendorf, USA) and reconstituted in 10 μl 0.1% trifluoroacetic acid. The peptides were desalted/concentrated using C18 ZipTip pipette tips (Millipore, USA) according to the manufacturer’s guidelines. The concentrated peptides were eluted with 0.8 μl matrix (10 mg/ml a-cyano-4-hydroxycinnamic acid prepared in 50% acetonitrile, 0.1% trifluoroacetic acid) and directly spotted onto a MALDI sample target plate.
e) Protein identification and bioinformatics analysis: Protein identification experiments were performed using ABSCIEX MALDI-TOF/TOF 5800 system. A streamline software, Protein Pilot (ABSCIEX, USA) was used to analyse the peak data with MASCOT. Enzyme of trypsin, 1 missed cleavage, fixed modifications of carbamidomethyl (C), variable modifications of oxidation (M), peptide mass tolerance (50 ppm), fragment mass tolerance (± 0.2 Da), peptide charge of 1+, and the monoisotopic were the parameters for searching. Only significant hits were accepted, as defined by the MASCOT probability analysis (p<0.05). The accession numbers of the identified proteins were uploaded into the Protein Analysis Through Evolutionary Relationships (PANTHER, http://pantherdb.org/) software. The default settings were used to generate the protein classifications.
Statistics
Values obtained in Figure 1 were assessed by unpaired, two tailed, equal variance Student’s t-test. Significant differences are shown by single symbols (*) indicating p<0.01. Error bars in figure represent SEM.
Results
ATP levels are lower in the absence of SIRT4 in mouse brain extracts
SIRT4 is known to increase the mitochondrial function [9]. Measuring the ATP levels or ATP formation is a way to measure mitochondrial function/activity. ATP levels in the wild type or SIRT4 KO mouse brain extracts (6 mice per group) were measured via a luminescence kit (Figure 1). ATP levels in wild type mouse brain extracts are higher than SIRT4 KO mouse brain extracts. This shows that the absence of SIRT4 caused a decrease in ATP levels indicating that SIRT4 might have a role in enhancing ATP formation. SIRT4 might activate one of the complexes in ATP production system such as protein complexes in electron transport chain.
Identification of SIRT4 interacting proteins
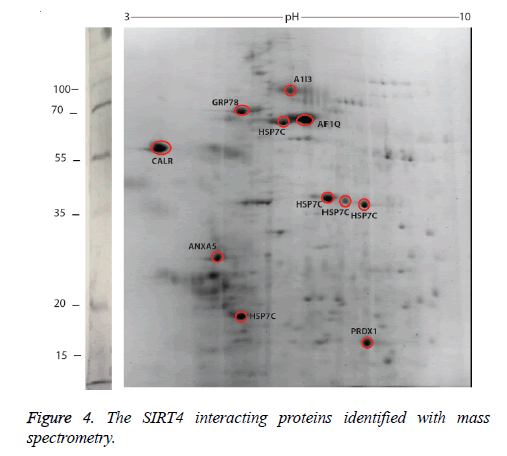
We identified the proteins that interact with SIRT4 and Figure 2 shows the experimental procedure. We have transfected MEFs with SIRT4-FLAG plasmid, immunoprecipitated SIRT4 interacting proteins using anti-FLAG M2 resin and run the eluate on 2D gel (Figures 2 and 3B). As a control experiment, RIPA buffer was run through a second column of anti-FLAG M2 resin instead of cell extract and the eluate was also run 2D gel (Figure 3A). As expected, the protein spots of the control eluate (Figure 3A) are quite different from the protein spots of the experimental eluate (Figure 3B). The protein spots of the experimental eluate (Figure 3B) were then identified via mass spectrometry (MALDI-TOF) as the proteins interacting with SIRT4 (Figure 4).
The target proteins were found to be HSC71, calreticulin, GRP78 (78 kDa glucose-regulated protein), annexin A5 and peroxiredoxin 1. We also identified alpha-1 inhibitor 3 and protein AF1q with low scores. A few of the aforementioned proteins are chaperones, or play roles in Ca+2 balance or redox regulation. They are all associated with mitochondria functions and stress response.
HSC71 is a molecular chaperone that plays roles in many cellular processes such as protecting the proteome from stress, folding of the newly synthesized polypeptide, the formation and destruction of the protein complexes [10]. It is a member of the heat shock protein 70 family and facilitates the proper folding of newly translated and misfolded proteins, as well as stabilize or degrade mutant proteins.
Calreticulin is a protein that functions in Ca+2 binding and storage and widely distributed in eukaryotic cells [11]. It was first described in the endoplasmic reticulum but also identified in many other cellular structures like the cytoplasm, the cell membrane, and the extracellular matrix. Calreticulin is involved in maintaining adequate calcium levels in organisms. It also functions as a chaperone to help other proteins fold correctly. GRP78 helps multimeric proteins to form in the endoplasmic reticulum [12]. It also helps with the correct folding of proteins and destruction of the misfolded proteins via interacting with DNAJC10.
Annexin A5 is an anti-coagulant protein, which is an indirect inhibitor of thromboplastin-specific complex that functions in the blood coagulation [13]. It is a cellular protein in the annexin group and commonly used to detect apoptotic cells by its ability to bind to phosphatidylserine [14].
Peroxiredoxin 1 plays role in the redox balance and function and decreases peroxides via thioredoxin. It is a member of the peroxiredoxin family of antioxidant enzymes, which reduce hydrogen peroxide and alkyl hydroperoxides [15]. It was also shown to have a proliferative effect and also takes place in cancer development or progression [16].
Alpha-1 inhibitor 3 is a protein that has a score below 100 in the list of target proteins. It is a protease inhibitor that binds to proteins via its thioester function [17,18]. Protein AF1q is another protein with a score below 100 among target proteins. It is the cofactor of the transcription factor TCF7 and involved in regulation of lymphoid development by driving multipotent hematopoietic progenitor cells towards a T cell fate [19].
Discussion and Conclusion
According to the mass spectrometry analysis, two of the proteins that interact with SIRT4 are HSC71 and GRP78 are chaperones. Chaperones function in many cellular processes including the proteolysis of the misfolded proteins, the formation and degradation of protein complexes, the folding of the newly synthesized protein and protecting the proteome from stress. They are located in different cellular compartments such as mitochondria, endoplasmic reticulum and cytoplasm. ATP binding and hydrolysis are essential in vitro and in vivo for the chaperone activity. The chaperones of the mitochondrial matrix form a complex interdependent chaperone network that is essential for most reactions of mitochondrial protein biogenesis.
Calreticulin is a Ca+2-binding and storage protein located in the endoplasmic reticulum lumen. For mitochondria to receive Ca+2 fast and adequately, the endoplasmic reticulum and mitochondria function closely with the help of Voltage-Gated Anion Channels (VDACs) surrounded by chaperones such as calreticulin and cannexin for buffering Ca+2 [20]. Another study showed that calreticulun is expressed in mitochondria and affects the function of ATP-dependent K-channels [21]. Peroxiredoxins regulate redox metabolism and H2O2 amount in cells by reducing peroxides and are expressed in mitochondria.
The proteins that interact with SIRT4 are generally ATP-dependent chaperones, regulate Ca+2 and redox metabolism and function in protein formation and folding. This is in line with our finding in this study that ATP production in SIRT4 KO mouse brain tissue is less than wild type. It is also in line with previous findings that SIRT4 has a role in clearing out the excess glutamate in synapses, which is related to Ca+2 and redox metabolism. In addition, chaperones are known to function in the regulation of neurotransmitter exocytosis [22]. SIRT4 was previously shown to play a role in glutamate-uptake [5]. Having found the chaperones as interacting partners of SIRT4, we now think that SIRT4 may also regulate neurotransmitter release via chaperones.
In future studies, different roles of SIRT4 will be elucidated by investigating the functions of SIRT4-interacting proteins and the related pathways. By this way, we will better understand the function of SIRT4 in mammalian brain and human diseases.
Funding
This work was supported by The Scientific and Technological Research Council of Turkey (TÜBITAK-115C098).
References
- Donmez G. The neurobiology of sirtuins and their role in neurodegeneration. TIPS 2012; 33: 494-501.
- Newman JC, He W, Verdin E. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem 2012; 287: 42436-42443.
- Haigis MC, Guarente LP. Mammalian sirtuins-emerging roles in physiology, aging, and calorie restriction. Genes Dev 2006; 20: 2913-2921.
- Komlos D, Mann KD, Zhuo Y, Ricupero CL, Hart RP, Liu AY-C, Firestein BL Glutamate dehydrogenase 1 and SIRT4 regulate glial development. Glia 2013; 61: 394-408.
- Shih J, Liu L, Mason A, Higashimori H, Donmez G. Loss of SIRT4 decreases GLT-1-dependent glutamate uptake and increases sensitivity to kainic acid. J Neurochemistry 2014; 131: 573-581.
- Karaca M, Frigerio F, Maechler P. From pancreatic islets to central nervous system, the importance of glutamate dehydrogenase for the control of energy homeostasis. Neurochem Int 2011; 59: 510-517.
- Danbolt NC. Glutamate uptake. Prog Neurobiol 2001; 65: 1-105.
- Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron 1998; 1: 623-634.
- Lombard DB, Tishkoff DX, Bao J. Mitochondrial sirtuins in the regulation of mitochondrial activity and metabolic adaptation. Handb Exp Pharmacol 2011; 206: 163-188.
- Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 2005; 62: 670-684.
- Gold LI, Eggleton P, Sweetwyne MT, Van Duyn LB, Greives MR, Naylor SM, Michalak M, Murphy-Ullrich JE. Calreticulin: non-endoplasmic reticulum functions in physiology and disease. FASEB J 2010; 24: 665-683.
- Wang M, Wey S, Zhang Y, Ye R, Lee AS. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid Redox Signal 2009; 11: 2307-2316.
- Meers P, Mealy T. Phospholipid determinants for annexin V binding sites and the role of tryptophan. Biochemistry 1994; 33: 5829-5837.
- Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pals ST, van Oers MH. Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 1994; 84: 1415-1420.
- Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H(2)O(2) accumulation for cell signaling. Cell 2010; 140: 517-528.
- Han YH, Kwon T, Kim SU, Ha HL, Lee TH, Kim JM, Jo EK, Kim BY, Yoon DY, Yu DY. Peroxiredoxin I deficiency attenuates phagocytic capacity of macrophage in clearance of the red blood cells damaged by oxidative stress. BMB Rep 2012; 45: 560-564.
- Wassler M, Esnard F, Fries E. Posttranslational folding of alpha-1-inhibitor 3. Evidence for a compaction process. JBC 1995; 270: 24598-24603.
- Esnard F, Gauthier F Maurizot JC. The interaction between rat plasma alpha 1 inhibitor 3 and chymotrypsin. A protease-protease inhibitor system which gives partially active complexes. Biochimie 1981; 63: 767-774.
- Park KW, Kim GE, Morales R, Moda F, Moreno-Gonzalez I, Concha-Marambio L, Lee AS, Hetz C, Soto C. The endoplasmic reticulum chaperone GRP78/BiP modulates prion propagation in vitro and in vivo. Sci Rep 2017; 7.
- Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signaling. Nat Rev Mol Cell Biol 2012; 13: 566-578.
- Shigaeva MI, Talanov EY, Venediktova NI, Murzaeva SV, Mironova GD. A role for calreticulin in functioning of mitochondrial ATP-dependent potassium channel. Biofizika 2014; 59: 887-894.
- Zinsmaier KE, Bronk P. Molecular chaperones and the regulation of neurotransmitter exocytosis. Biochem Pharmacol 2001; 62: 1-11.