Research Article - Archives of General Internal Medicine (2023) Volume 7, Issue 6
Genetic Variants Affecting Insulin Signaling Pathways and Risk of Diabetes: A Comprehensive Systematic Review
Namra Vinay Gohil1, Vaishnavi Kandukuri2, Navya Pillikunte Doddareddy3, Poornima Jaiswal Charpuria4, Shresta Mary Kurian5, Vishva Babu6, Narendranath Reddy Ganampet7, Mihirkumar P.Parmar8*, Vishal Venugopal91Department of Internal Medicine, Medical College Baroda, Vadodara, India
2Department of Internal Medicine, Gandhi Medical College and Hospital, Hyderabad, India
3Department of Internal Medicine,Bangalore Medical College and Research institute, Bangalore, India
4Department of Internal Medicine,Osmania Medical College, , Hyderabad, India
5Department of Internal Medicine, Chalmeda Anand Rao Institute of Medical Sciences, Hyderabad, India
6Department of Internal Medicine,Jawaharlal Institute of Postgraduate Medical Education and Research, Pondicherry, India
7Department of Internal Medicine,Kamineni Academy of Medical Sciences and Research Centre, Hyderabad, India
8Department of Internal Medicine, Gujarat Medical Education and Research Society, Vadnagar, India
9Department of Internal Medicine, Bhaarath Medical College and Hospital, Chennai, India
- *Corresponding Author:
- Mihirkumar P. Parmar
Discipline of Pharmacology, School of Pharmacy
University of the Western Cape
Bellville 7535, South Africa
Tel: +27219593229/2190
Fax: +278615107002
E-mail: mmihir981@gmail.com
Received: 23-Nov-2023, Manuscript No. AAAGIM-23-119276; Editor assigned: 27- Nov -2023, PreQC No. AAAGIM-23-119276(PQ); Reviewed: 12-Dec-2023, QC No. AAAGIM-23-119276; Revised: 15- Dec -2023, Manuscript No. AAAGIM-23-119276(R); Published: 22- Dec-2023, DOI:10.35841/aaagim-7.6.202
Citation: Gohil N.V, Kandukuri V, et al. Genetic variants affecting insulin signaling pathways and risk of diabetes: A comprehensive systematic review. Arch Gen Intern Med. 2023;7(6):202
Abstract
Purpose: The complicated condition known as type 2 diabetes mellitus (T2DM) has a diverse genetic and environment-related cause. MIM125853 is among the heterogeneous diseases that can also be characterized as a multifactorial disorder of glucose metabolism with genetic susceptibility of an anomaly in the insulin pathway. We conducted the review with aim to assess the genetic variants affecting the insulin pathway as well as evaluate the risk of diabetes in association with genetic variants. Methods: We used literature of 13 researches that met the inclusion criteria after the process of selection and analyzation. Our analysis included 59,593 participants in total with a gender distribution of 48% males (n=24,591) and 52% females (n=35,002). Result: Gene variants such as PPARG, SLC30A8, KCNJ11, TCF7L2 and many others plays a pivotal role in the development of type-II diabetes as well as optimal functioning of insulin metabolic pathway for glucose metabolism and distribution with circulation. Conclusion: KCNJ11 regulates insulin release in collaboration with other genes including ABCC8, KAPN10, IRS1 and TCF7L2. Reduced mutual expression of these kinds of genes could contribute to DM susceptibility. However, it is still unknown how precisely the combination of these genes’ functions in the control of insulin secretion.
Keywords
PPARG, SLC30A8, KCNJ11, TCF7L2, diabetes mellitus type- II, insulin signaling pathway.
Introduction
Numerous diseases have been identified as polygenic disorders with certain allele variants alleviating or doubling the development and progression of the disease [1]. The multifactorial diseases are considered as contributory factors to several secondary diseases as well as affect the quality of life immensely [2]. The genetic variants linked to diseases are a major concern for scientists in order to get hold of diseases as well as utilize the genetic findings for future generations. The variation of genetic alleles identified and marked for association with any particular disease can be detected in people to depict the risk of developing the disease or extent of progression of an already existing condition [3]. Etiological studies utilize genes to determine the root cause as well as take it into account while projecting results. Genetic studies of any extent involve the variant study, polymorphism of genes and genetic mapping of DNA with mutations of alleles compared to their original genetic makeup [4].
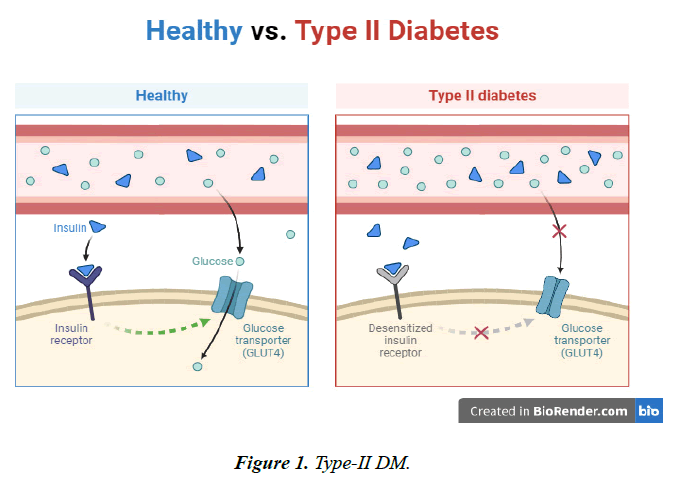
Diabetes Mellitus (MIM125853) is among the heterogeneous diseases that can also be characterized as a multifactorial disorder of glucose metabolism [5]. It can mainly be described as a defect by an impaired inulin response to the desired level as well as insufficient production of insulin by the pancreatic β cells due to its attempt to decrease insulin resistance in cells as well as normalize the circulatory sugar levels [6]. Diabetes mellitus is also marked as a metabolic disorder with the inability of body to perform optimal carbohydrate metabolism at the cellular level as well as causing chronic hyperglycemia in the body. Chronic hyperglycemia is linked to the development of obesity, PCOS, dyslipidemia as well as secondary diseases of diabetes such as nephropathy, retinopathy and cardiovascular diseases [5].
There has been an increase in the prevalence of diabetes mellitus worldwide in the past decade owing to several genetic and environmental factors. 463 million adults are reported to be suffering from diabetes worldwide with almost 90% of individuals suffering from type-2 diabetes mellitus [7]. In the year 2019, 4.2 million people worldwide died from diabetes, which affected a total of 466 million individuals between the ages of 20 and 79, based to the 9th edition of the International Diabetes Federation (IDF) Atlas. In the year 2045, 700 million people are anticipated to have this debilitating illness [8].
Several studies have been conducted with the aim to identify the cause of the disease mainly the root cause in the genetic makeup for progression and development of disease. It has also been under great focus to determine the association of genetic variants to diabetes etiology and biological mechanism of insulin pathway as well as signaling and functioning. Several diabetogens with great impact on insulin receptors, insulin pathways as well as glucokinase genetic makeup are studied [5].
Diabetes mellitus has been subjected to genetic studies to determine the cause as well as identification of f scription genes responsible for the encoding of HNF1 a HNF 4 a and B (hepatocyte nuclear factor) and IPF 1 (insulin promoter factor) along with mitochondrial mutation in DNA are studied. Analysis for investigation of diabetes Mellitus genetic makeup require researches by utilizing high density DNA chips, complete genome DNA methylation analysis as well as sequence of entire genome [9]. Mainly, TCF7L2 (T at rs7903146), PPARG (Pro12), KCNJ11 (Lys23) have been concerned with diabetes mellitus type 2 [10]. PPARG has been associated significantly with Asians when compared to Europeans and elevated relationship among North Europe rather than Southern region of Europe [11].
Obesity is an important risk indicator for T2D, and many individuals with a genetic susceptibility to the disease additionally exhibit an inclination to weight gain [3]. Several potential genes have been linked to T2D [4–7], but many of the discoveries have been challenging to confirm. A small number of genes, such as those expressing PPARG, calpain 10, Kir 6.2, and the insulin receptor substrate 1 (IRS1), have received support in in-depth meta-analyses. The PPARG P12A mutation improves the responsiveness to insulin and prevents type 2 diabetes. The population-attributable risk of the prevalent allele is roughly 25%, despite the fact that carriers of the uncommon A allele have a 15% reduction in their personal risk. Introns 43 and 44 of the genomes encoding the cystein protease calpain 10 (CAPN10) confer higher vulnerability to developing insulin resistance and type 2 diabetes (T2D). Combined with the sulfonylurea receptor SUR1 (ABCC8), the ATP-sensitive potassium channel Kir 6.2 (KCNJ11) forms an octamer, which is a polypeptide that controls the potential for transmembrane transport and, in turn, glucose-stimulated insulin release in pancreatic β cells. T2D has been linked to a KCNJ11 E23K polymorphism. It has been demonstrated that individuals who carry the IRS1 gene's G972R polymorphism have less insulin in their islets of the pancreas. Although the G972R polymorphism may have a function in T2D, two recent, sizable case-control investigations were unable to confirm this connection [12].
A study conducted to determine the effect of PPARG and KCNJ11 on development of type two diabetes mellitus employed 6447 research participants including diabetic and glucose tolerant subjects. In the two independent evaluations type 2 diabetes was significantly linked with the K allele of the KCNJ11 Glu23Lys (odds ratio, 1.19) but not with the PPARG Pro12Ala. There was no indication of a mutually beneficial relationship between the two SNPs, according to the integrated analysis, which showed that they raised the likelihood of type 2 diabetes in a mutually reinforcing way. The hazard ratio for type 2 diabetes increased incorporating the presence of the risk variant by 1.14, according to the evaluation of a model with equivalent additive impacts from the two variants (P 0.003). Two SNPs combined to produce a 28% population-attributable chance of developing type 2 diabetes. The KCNJ11 Glu23Lys and PPARG Pro12Ala polymorphisms may raise the likelihood of the development of type 2 diabetes in a complementary manner, however the findings did not support an additive relationship between them [4].
Aims & Objectives
• To assess the genetic variants affecting insulin pathway
• To evaluate the risk of diabetes in association with genetic variants
• Provide emerging medical professionals with a better view on the best ways to manage and treat disease
Materials and Methods
Study Design
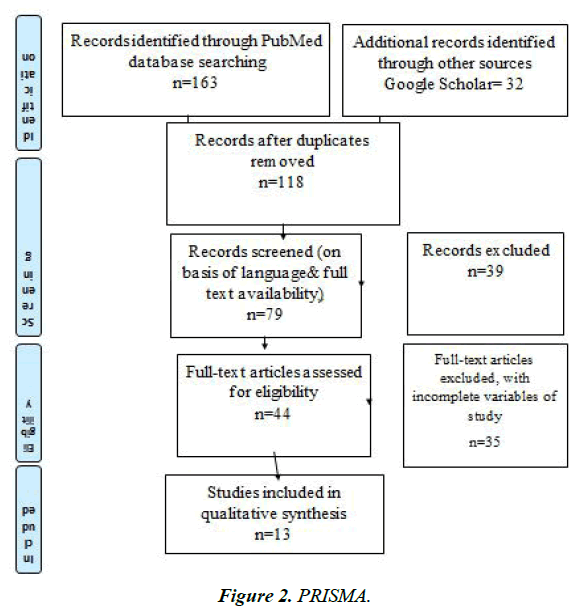
To assess the elevated likelihood of diabetes from genetic variations, this review is based on the literature and includes papers from research. Moreover, due to the distinctive features of the study we examined and the lack of data on the researched topic. We included cohort and observational studies along with genetics reports that reported the risk of diabetes mellitus type 2 related to genetic variants because they are more probable in illustrating the relational effects of any event as a result. The Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) method was used to review the literature.
Inclusion Criteria
• Studies that investigate the association between genetic variants and risk of diabetes (both type 1 and type 2).
• Studies that focus on genes directly involved in insulin signaling pathways, such as those related to insulin receptor, IRS, PI3K, Akt, etc.
• Human studies, including case-control, cohort, and population-based studies.
• Studies that provide information on the specific genetic variants, alleles, or mutations.
• Published studies in peer-reviewed journals.
• Studies published in the English language.
Exclusion Criteria:
• Animal studies or in vitro studies.
• Studies that do not focus on genetic variants within insulin signaling pathways.
• Studies with inadequate information on genetic variants or risk estimates.
• Review articles, case reports, conference abstracts, and editorials.
• Non-English language studies.
• Studies that do not report primary outcomes related to diabetes risk.
Search strategy
The literature-based systematic review for the most recent research employed the PRISMA strategy. The search method developed suggested including publications on genetic variants in relation to insulin pathway in diabetes mellitus. The terms genetic variants, Insulin pathway susceptibility, insulin signaling, Diabetes mellitus gene mutations, risk of diabetes mellitus and variants were utilized in an electronic search on PubMed Online, EBSCO and Scopus along with google scholar. To determine if every component met the criteria for inclusion, it was thoroughly reviewed and analyzed. It was evaluated and examined. If they satisfied all inclusion criteria, they were simply included in the research.
Results and Discussion
The research studies selected were utilized to obtain data about genetic variation and their linkage to the development and occurrence via an effect on insulin signaling pathways. A literature-based analysis was conducted that included 13 researches included after the process of selection and analyzation. The characteristics of research studies are elaborated in Table 1. Our analysis included 59,593 participants in total with a gender distribution of 48% males (n=24,591) and 52% females (n=35,002).
| Study | Randomization Process | Deviation from Intervention | Missing outcome data | Measurement of Outcome | Selection of Reported Result | Overall, Bias Risk |
|---|---|---|---|---|---|---|
| D1 | D2 | D3 | D4 | D5 | Results | |
| Weedon et al. [13] | Low | Low | Low | Low | Low | Low risk |
| Hansen et al. [4] | Low | Low | Low | Low | Low | Low risk |
| Lyssenko et al. [12] | Low | Low | Low | Low | Low | Low risk |
| Uma Jyothi et al. [14] | Low | Low | Low | Low | Some Concerns | Some Concerns |
| Gloyn et al. [15] | Low | Low | Low | Low | Low | Low risk |
| Nielsen et al. [16] | Low | Low | Low | Low | Low | Low risk |
| Zhang et al. [17] | Low | Low | Low | Low | Low | Low risk |
| Scott et al. [18] | Low | Low | Low | Some Concerns | Some Concerns | Some Concerns |
| Weedon et al. [19] | Low | Low | Low | Low | Some Concerns | Some Concerns |
| Zeggini et al. [20] | Low | Low | Some Concerns | Low | Low | Some Concerns |
| Drake et al. [21] | Low | Low | Low | Low | Low | Low risk |
| Cropano et al. [22] | Low | Low | Low | Some Concerns | Some Concerns | Low risk |
| Mashal et al. [23] | Low | Low | Low | Low | Low | Low risk |
Table 1. Bias Assessment of research studies via Cochrane Risk Assessment Tool.
Bias among the researchers was identified by using the Cochrane Risk assessment tool to devise optimal results and utilization of low bias risk data. There were nine studies with low bias risk whereas the rest of the four study had some concerns for the data reported in the studies. The Bias tool result is mentioned in Table 2. The total studies were from various geographical regions to include diversity and procure an exceptional outcome for future prospects related to gene variants and diabetes mellitus 2 with respect to insulin pathways. The studies were conducted in the United Kingdom, Sweden, Denmark, India, Finland as well as United States of America and Jordan.
| Study | Year | Location | Sample size | Age | Gender M/F | Method Used | Variants & OR | 95% CI (Range) | |
|---|---|---|---|---|---|---|---|---|---|
| Weedon et al. [13] | 2006 | UK | 6077 | 40.25 | 3231/ 2846 | Genotyping using TaqMan-based assay | Glu23Lys KCNJ11 | 1.14 | 1.05-1.23 |
| Pro12Ala PPARG | 1.29 | 1.14- 1.45 | |||||||
| TCF7L2 rs7903146 | 1.48 | 1.36-1.60 | |||||||
| Hansen et al. [4] | 2005 | Denmark | 7432 | 52.34 | 4285/ 3138 | chip-based, matrix-assisted laser desorption of flight mass spectrometry analysis of PCR-generated primer extension | K allele of the KCNJ11 Glu23Lys | 1.19 | 1.09-1.30 |
| Pro12Ala | _ | ||||||||
| Lyssenko et al. [12] | 2005 | Finland | 2293 | 45.1 | 1051/ 1242 | Stored in EDTA-coated vacutainer and genotypes as 15-SNP panel | PPARG P12A | 1.7 | 1.1-2.7 |
| CAPN10 SNP43G | 1.5 | 1.0-2.2 | |||||||
| UCP2 866G | 1.4 | 1.0-1.9 | |||||||
| IRS1 G972R | 1.3 | 0.8-2.1 | |||||||
| Uma Jyothi et al. [14] | 2014 | India | 1379 | IRS1 | 1.8 | 1.13- 2.78 | |||
| CAPN10 | 1.33 | 1.07-1.68 | |||||||
| PPARG | 0.99 | 0.91-2.07 | |||||||
| Gloyn et al. [15] | 2002 | UK | 2486 | 978/ | E23K of KCNJ11 | 1.18 | 1.04-1.34 | ||
| 1508 | SUR1 of ABCC8 | 1.04 | 0.91-1.18 | ||||||
| Nielsen et al. [16] | 2003 | 519 | 57.1 | 242/ 277 | Kir6.2 of KCNJ11 | 1.49 | 1.20-1.83 | ||
| Zhang et al. [17] | 2006 | 3520 | 56.6 | 886/ 687 | TCF7L2 | 2.15 | 1.48- 3.13 | ||
| s12255372 | |||||||||
| Scott et al. [18] | 2006 | Finland | 1749 | 55.1 | 966/ | Sequenom homogeneous MassEXTEND assay. | TCF7L2 | 1.36 | 1.15- 1.61 |
| 783 | s12255372 | ||||||||
| TCF7L2 | 1.33 | 1.14-1.56 | |||||||
| s7903146 | |||||||||
| Weedon et al. [19] | 2004 | UK | 4148 | 56 | 2311/ 1837 | HNF4a P2 rs4810424 | 1.29 | 0.96- 1.73 | |
| HNF4a P2 | 1.32 | 1.00- 1.75 | |||||||
| rs2144908 | |||||||||
| Zeggini et al. [20] | 2005 | UK | 2228 | Pro12Ala PPARG | 1.4 | 1.12- 1.76 | |||
| Drake et al. [21] | 2017 | Sweden | 26132 | 69.1 | 9930/ 16202 | Sequenom MassARRAY iPLEX | SLC30A8 CC | 1.16 | 1.07- 1.24 |
| Cropano et al. [22] | 2017 | USA | 955 | 13.3 | 394/ 561 | matrix assisted–based laser desorption-ionization time-of-flight mass spectrometry on the MassARRAY platform (Sequenom) | TCF7L2 | 2.24 | 1.37- 3.61 |
| s7903146 | |||||||||
| Mashal et al. [23] | 2020 | Jordan | 684 | 58.2 | 314/ 370 | DNA was extracted using the Wizard genomic purification kit | SLC30A8 CC | 2.44 | 1.16- 5.12 |
| rs 1326634 | |||||||||
Table 2. Characteristics of Research Studies.
KCNJ11
KCNJ11 stands for Potassium inwardly, rectifying channel subfamily J member 11, which is present in mammalian cells within potassium channels responsible for encoding the pivotal membrane protein as well as the potassium channel of the inward rectifier. It is responsible for the production of ATP-sensitive potassium channel subunits. Variation or defect in this gene has been associated with non-insulindependent diabetes mellitus type II (NIDDM) also called as type 2 diabetes mellitus [24]. We included research studies employing the KCJN11 gene with risk ratio of development of diabetes mellitus 2. The variant of Glu23Lys KCNJ11 in the study was 1.14 95% (CI1.05-1.23) among research participants of the United Kingdom (11) whereas it was 1.19 95% (CI 1.09-1.30) in Denmark by the study [25].
Kir6.2
The ATP-sensitive potassium (KATP) channel in pancreatic beta cells mediates insulin secretion. The channel itself is a heteromeric polypeptide made up of sulfonylurea receptor 1 subunits around the pore and four inward-rectifier potassium ion channel (Kir6.2) tetramers that assemble into the KATP channel's gate. The potassium channel gene KCNJ11, which belongs to the subgroup J of potassium channel genes, encodes the protein Kir6.2. Many research investigations have documented how the KCNJ11 gene's single-molecule polymorphisms and their interactions with one another affect DM predisposition [24]. Kir6.2 of KCNJ11 reported the odds ratio of developing diabetes mellitus as 1.49 (CI 1.20-1.83) which is also necessary for the metabolic pathway of insulin production, signaling and function [16].
In a variety of organs and cell types, including beta-cells in the pancreas, the brain, kidneys, heart, skeletal, and smooth muscle mass, K ATP channels control the electrically generated signals of the plasma cell membrane [26].
PPARG
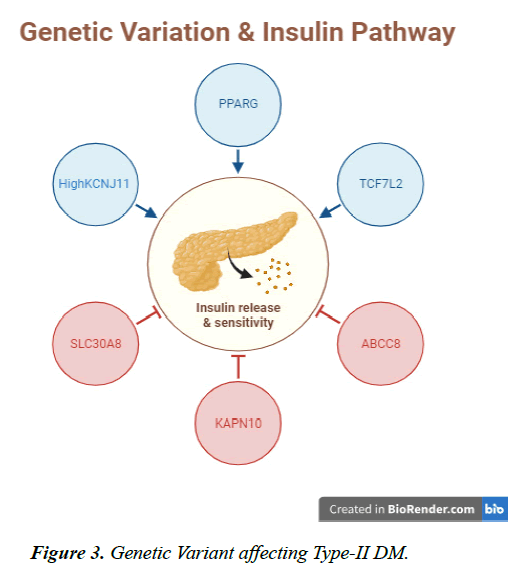
PPARG stands for Peroxisome proliferator-activated receptors are ligand-activated gene transcription variables that belong to the nuclear receptor giant family [27]. The additional mRNA splicing process that produces PPAR-2 controls the transcription and expression of several genes of interest. The genes in question have been demonstrated to play an essential part in atherosclerosis, the metabolism of glucose and lipids, and adipocyte formation [28] (Figure.1- 3).
SLC30A8
Recent genome-wide association research investigations have shown that the solute carrier family 30-member 8 gene (SLC30A8)'s prevalent variants enhance type 2 diabetes susceptibility. Zinc transporter-8 (ZnT8), which is encoded by SLC30A8, transports zinc ions from the cytoplasm to insulin granulates. Despite the high zinc content of insulin granules is widely recognized, its physiological significance is yet unknown. Studies are being done to determine the functional relationship amongst Slc30a8 deletion and hepatic insulin clearance as well as cell-specific Slc30a8 deficiency [29].
SLC30A8 stands for solute carrier family 30 member 8 which encodes the zinc efflux transporter that administer the reservation of zinc in the intracellular vesicles. This gene is shows prevalent expression in the pancreas primarily β- cells or Islet of Langerhans in the pancreas. It also works synergistically with Insulin secretin cells-1 or INS cells that in turn affect with the development of progression of diabetes mellitus type2 or insulin independent diabetes. It is also known as ZNT8 or ZnT-8 showing the role of zinc in this genetic expression [30].
ZnT8 is required for the crystallization of insulin molecules and effective insulin absorption in insulin granules, according to recent investigations of Slc30a8 in mice, however the precise contribution of ZnT8 to type 2 diabetes susceptibility remains unclear [31]. The research also showed increased risk of diabetes development associated with this gene variant i.e., SLC30A8 CC with OR of 2.44 (CI 1.16-5.12) [23].
TCF7L2
TCF7L2 is the abbreviation of transcription factor 7 like 2 also known as TCF-4. This particular gene expresses a transcription factor with a high mobility group (HMG) box that is essential for the insulin activation pathway. The protein was recently linked to the regulation of blood sugar levels. This gene's variation in genes is linked to a higher chance of developing type 2 diabetes [32]. The primary defects leading T2D to develop are insulin resistance in the liver and cellular dysfunctional behavior. Therefore, hepatic insulin sensitivity (SI) is essential in contributing to the development of diabetes [33]. It has been subjected that single nucleotide polymorphism (SNP) in the TCF7L2 gene region encodes Wnt pathway, which activates β-cells of the pancreas as well as other lineages of cells responsible for glucose metabolization and production [34]. The research conducted in America elucidated OR of 2.24 (1.37-3.61) with TCF7L2 variation depicting an increased risk of DM-II development [22].
The study conducted to determine the association of TCF7L2 risk allele 7903146 elucidated two times elevated risk of glucose associated metabolic disease among Caucasians with similar trend observed in African Americans and Hispanic as well. It had been related to the development of endogenous suppression of glucose production as well as reduced β cell responsiveness to the cellular signaling. This result indicates increase risk of diabetes development among adolescents with impaired insulin pathway [22].
ABCC8
ABCC8 can be recognized as ATP binding cassette subfamily C member 8 with other names of TNDM2, SUR1 and PNDM3 as well. The ATP-binding cassette (ABC) transporter superfamily includes the protein that this gene encodes. Multiple substances are transported across extracellular and inner cell membranes by ABC proteins. The seven different subdivisions of ABC genes are ABC1, ALD, OABP, GCN20, MDR/TAP, MRP, and White [35]. This protein controls the discharge of insulin and ATP-sensitive channels that release potassium. Patients with hyperinsulinemic hypoglycemic of infancy, an autosomal recessive condition of uncontrolled and excessive insulin production, have been indicated to have polymorphisms in the ABCC8 gene and abnormalities in the protein that was encoded. Additionally, diabetes mellitus type II, a condition that is autosomal dominant and characterized by impaired insulin production, has been linked to abnormalities [36].
A study conducted to determine the association of the ABCC8 gene with the SUR1 subunit identified mutations within the gene and reported that ABCC8 mutation can act as a causative factor in the development of permanent (PNDM) or transient neonatal diabetes mellitus (TNDM) as well as type-II diabetes with permanent phenotypic presentation. The observation that many newborn diabetes patients with ABCC8 mutations are KCNJ11-negative supports the idea suggesting the interactions between Kir6.2 and SUR1 are essential for proper channel activity and, consequently, for optimum production of insulin [37].
CAPN10
CAPN 10 with the full name of calpain 10 is also called NIDDM1 as they are a family of cysteine proteases dependent on calcium. A substantial subunit is encoded by this gene. It is an unusual calpain because it does not have the calciumbinding region that is similar to the calmodulin and has a diverged C-terminal domain instead. It is structured similarly to Caplains 5 and 6. This gene is found in the NIDDM1 locus and is linked with type 2 or non-insulin-dependent diabetic mellitus (NIDDM) [38]. The CAPN10 in the study was reported to have OR of 1.33 (CI1.07- 1.68) [14].
Numerous physiological functions, including cell communication, a process called apoptosis and exocytosis, the metabolic process of mitochondria, and cytoskeletal reshaping have been linked to calpain activity which plays a role in insulin resistance [39].
IRS
Insulin Receptor substrate 1gene which is a crucial component of the insulin-signaling routes, and it has been suggested that variations in the IRS1 gene influence a person's propensity for developing type 2 diabetes. Linkage study has determined that the IRS1 locus on 971Arg allele shows 50% decreased insulin sensitivity [40].
The variation in the transcription of IRS1 in adipocytes and skeletal muscle and the correlation of IRS1 polymorphisms with the development of type 2 diabetes and related phenotypes indicate its role for this gene in the pathophysiology of type 2 diabetes [41]. IRS1 gene for DM-II with OR 1.30 and CI 0.83- 2.16 indicated increased risk of this gene with DM II.
HNF4α P2
Hepatocyte nuclear factor 4 alpha is also known as MODY-1 and TCF. This gene produces the nuclear transcription factor, a protein that engages DNA as a homodimer. Monogenic autosomal dominant non-insulin-dependent diabetes mellitus type I has been found linked to polymorphisms in this gene [42].
Fasting causes an acute induction of this isoform, while type two diabetes (T2D) causes a constant increase. The overexpression of TET3 induced by glucagon, which had not previously been demonstrated to have a role in HGP, results in the production of the P2 isoform. By bringing TET3 to the P2 promoter region, FOXA2 causes the promoter itself to be demethylated, which boosts transcription. This mechanism can act as a layout for targeting the genetic therapies for type- II DM [43].
There are many other factors that might act as contributory factors for development or progression of type-II diabetes mellitus. Obesity and overweight along with systemic inflammation have been linked to increased risk of diabetes development. Sedentary lifestyle and environmental stimuli along with disease associated inflammation causing insulin resistance can act as causative agents. Hormonal changes such as PCOS as well as diagnosis of gestational diabetes mellitus (GDM) increase the risk of developing diabetes mellitus many folds.
Conclusion
Several of the most prevalent illnesses in the world, DM, have significant social and financial costs. The KATP channel may work in part thanks to Kir6.2, HNFα and IRS 10. Several prominent variants in this gene's region can interfere with the activation and signaling pathway of insulin, which in turn lowers the KATP channel's potential and causes DM. The research makes clear that a number with KCNJ11 gene variations are connected to various forms of DM.
KCNJ11 regulates insulin release in collaboration with other genes including ABCC8, KAPN10, and TCF7L2. Reduced mutual expression of these kinds of genes could contribute to DM susceptibility. However, it is still unknown how precisely the combination of these genes’ functions in the control of insulin secretion. Future research is advised to determine the precise function of KCNj11 gene variations and how they interact with additional genes in DM in order to identify potential treatments and diagnose this widespread condition.
References
- Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385-9.
- Reich DE, Lander ES. On the allelic spectrum of human disease. TRENDS in Genetics. 2001;17(9):502-10.
- Yang Q, Khoury MJ, Botto L, et al. Improving the prediction of complex diseases by testing for multiple disease-susceptibility genes. Am J Hum Gene. 2003;72(3):636-49.
- Hansen SK, Nielsen EM, Ek J, et al. Analysis of separate and combined effects of common variation in KCNJ11 and PPARG on risk of type 2 diabetes. J Clin Endocrinol Metabolism. 2005;90(6):3629-37.
- Vionnet N, Dupont S, Gallina S, et al. Genomewide search for type 2 diabetes–susceptibility genes in French Whites: evidence for a novel susceptibility locus for early-onset diabetes on chromosome 3q27-qter and independent replication of a type 2–diabetes locus on chromosome 1q21–q24. Am J Hum Gene. 2000;67(6):1470-80.
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes care. 2010;33(Supplement_1):S62-9.
- 9th edition | IDF diabetes Atlas [internet]. https://diabetesatlas.org/atlas/ninth-edition Accessed 29 Aug 2023
- IDF. IDF Diabetes Atlas 2019. 9th edn (International Diabetes Federation, 2019). https://www.diabetesatlas.org/upload/resources/material/20200302_133351_IDFATLAS9e-final-web.pdf Accessed 30 Aug 2023
- Elbein SC. The genetics of human noninsulin-dependent (type 2) diabetes mellitus. The Journal of nutrition J Nutr. 1997;127(9):1891S-6S.
- Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nature genetics. 2000;26(1):76-80.
- Prudente S, Morini E, Trischitta V. Insulin signaling regulating genes: effect on T2DM and cardiovascular risk. Nat Rev Endocrinol. 2009;5(12):682-93.
- Lyssenko V, Almgren P, Anevski D, et al, Botnia Study Group. Genetic prediction of future type 2 diabetes. PLoS medicine. 2005;2(12):e345.
- Weedon MN, McCarthy MI, Hitman G, et al. Combining information from common type 2 diabetes risk polymorphisms improves disease prediction. PLoS medicine. 2006;3(10):e374.
- Uma Jyothi K, Jayaraj M, Subburaj KS, et al. Association of TCF7L2 gene polymorphisms with T2DM in the population of Hyderabad, India. PLoS One. 2013;8(4):e60212.
- Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic β-cell KATP channel subunits Kir6. 2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52(2):568-72.
- Nielsen EM, Hansen L, Carstensen B, et al. The E23K variant of Kir6. 2 associates with impaired post-OGTT serum insulin response and increased risk of type 2 diabetes. Diabetes. 2003;52(2):573-7.
- Zhang C, Qi L, Hunter DJ, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene and the risk of type 2 diabetes in large cohorts of US women and men. Diabetes. 2006;55(9):2645-8.
- Scott LJ, Bonnycastle LL, Willer CJ, et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. diabetes. 2006;55(9):2649-53.
- Weedon MN, Owen KR, Shields B, et al. Common variants of the hepatocyte nuclear factor-4α P2 promoter are associated with type 2 diabetes in the UK population. Diabetes. 2004;53(11):3002-6.
- Zeggini E, Parkinson JR, Halford S, et al. Examining the relationships between the Pro12Ala variant in PPARG and Type 2 diabetes‐related traits in UK samples. Diabetic medicine. 2005;22(12):1696-700.
- Drake I, Hindy G, Ericson U, et al. A prospective study of dietary and supplemental zinc intake and risk of type 2 diabetes depending on genetic variation in SLC30A8. Genes & nutrition. 2017;12:1-1.
- Cropano C, Santoro N, Groop L, et al. The rs7903146 variant in the TCF7L2 gene increases the risk of prediabetes/type 2 diabetes in obese adolescents by impairing β-cell function and hepatic insulin sensitivity. Diabetes care. 2017;40(8):1082-9.
- Mashal S, Khanfar M, Al-Khalayfa S, et al. SLC30A8 gene polymorphism rs13266634 associated with increased risk for developing type 2 diabetes mellitus in Jordanian population. Gene. 2021;768:145279.
- Haghvirdizadeh P, Mohamed Z, Abdullah NA, et al. KCNJ11: genetic polymorphisms and risk of diabetes mellitus. J Diabetes Res. 2015;2015.
- Shoily SS, Ahsan T, Fatema K, et al. Common genetic variants and pathways in diabetes and associated complications and vulnerability of populations with different ethnic origins. Scientific Reports. 2021;11(1):7504.
- Seino S, Miki T. Physiological and pathophysiological roles of ATP-sensitive K+ channels. Prog Biophys Mol Biol. 2003;81(2):133-76.
- Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645-50.
- Li AC, Glass CK. PPAR-and LXR-dependent pathways controlling lipid metabolism and the development of atherosclerosis. J Lipid Res. 2004;45(12):2161-73.
- Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331-6.
- Zeng Q, Tan B, Han F, et al. Association of solute carrier family 30 A8 zinc transporter gene variations with gestational diabetes mellitus risk in a Chinese population. Front Endocrinolog. 2023;14:1159714.
- Pound LD, Sarkar SA, Benninger RK, et al. Deletion of the mouse Slc30a8 gene encoding zinc transporter-8 results in impaired insulin secretion. Biochemical Journal. 2009;421(3):371-6.
- Amin M, del Bosque-Plata L, Gragnoli C. Novel linkage and association of TCF7L2 variants with PCOS in Italian families. Eur Rev Med Pharmacol Sci. 2023;27(15).
- Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881-5.
- Smith U. TCF7L2 and type 2 diabetes—we WNT to know. Diabetologia. 2007;50(1):5-7.
- Silvestri F, Tromba V, Schiaffini R, et al. The Arg1379His mutation in ABCC8 causes monogenic diabetes with variable phenotype presentation and incomplete penetrance. Acta Diabetologica. 2023;60(7):989-93.
- Tran NQ, Truong SD, Ma PT, et al. Association of KCNJ11 and ABCC8 single-nucleotide polymorphisms with type 2 diabetes mellitus in a Kinh Vietnamese population. Medicine. 2022;101(46).
- Patch AM, Flanagan SE, Boustred C, et al. Mutations in the ABCC8 gene encoding the SUR1 subunit of the KATP channel cause transient neonatal diabetes, permanent neonatal diabetes or permanent diabetes diagnosed outside the neonatal period. Diabetes, Obesity and Metabolism. 2007;9:28-39.
- Castro-Martínez AG, Sánchez-Corona J, Vázquez-Vargas AP, et al. Association analysis of calpain 10 gene variants/haplotypes with gestational diabetes mellitus among Mexican women. Cellular and Molecular Biology. 2018;64(3):81-6.
- Pánico P, Salazar AM, Burns AL, Ostrosky-Wegman Pet al. Role of calpain-10 in the development of diabetes mellitus and its complications. Arch Med Res. 2014;45(2):103-15.
- Celi FS, Negri C, Tanner K, et al. Molecular scanning for mutations in the insulin receptor substrate‐1 (IRS‐1) gene in Mexican Americans with Type 2 diabetes mellitus. Diabetes/metabolism research and reviews. 2000;16(5):370-7.
- Kovacs P, Hanson RL, Lee YH, et al. The role of insulin receptor substrate-1 gene (IRS1) in type 2 diabetes in Pima Indians. Diabetes. 2003;52(12):3005-9.
- Love-Gregory L, Permutt MA. HNF4A genetic variants: role in diabetes. Curr Opin Clin Nutr Metab Care. 2007;10(4):397-402.
- Li D, Cao T, Sun X, et al. Hepatic TET3 contributes to type-2 diabetes by inducing the HNF4α fetal isoform. Nat Com. 2020;11(1):342.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref