Research Article - Biomedical Research (2023) Volume 34, Issue 1
Genetic influences of major lung diseases on lung cancer: A systematic and bioinformatics approach.
Amina Rownaq1*, Shahinul Islam SM1, Md Nurul Haque Mollah2
1Plant Biotechnology and Genetic Engineering Lab, Institute of Biological Sciences, University of Rajshahi, Bangladesh
2Department of Statistics, Bioinformatics Lab, University of Rajshahi, Bangladesh
- Corresponding Author:
- Amina Rownaq
Plant Biotechnology and Genetic Engineering Lab
Institute of Biological Sciences
University of Rajshahi
Bangladesh
Accepted date: February 8, 2023
Abstract
Background: Cancer comorbidity with lung diseases is evident from epidemiological and clinical data. Fatality relationships analysis is tough to check intimately victimization by using conventional endocrinological strategies. We tend to developed tissue transcript method to determine genetic influences of lung diseases on lung cancer. Different lung diseases have significant association with the threat of lung cancer
Methods: By using two lung carcinoma microarray datasets and four lung diseases (Asthma, Bronchitis, COPD and COVID) microarray datasets, Differentially Expressed Genes (DEGs) were determined comparing normal and diseases-related lung tissue. The betweeness and interaction of DEGs was detected through the STRING tool in Cytoscape software. Gene Ontology (GO) and enriched molecular pathway analysis of DEGs were analyzed via DAVID software. We also constructed a DEG-TF and DEG-miRNA interactions networks for analysis of the gene targets of Transcription Factors (TFs) and microRNA (miRNAs) through miRNet. For survival analysis we used online SurvExpress database.
Results: One hundred nineteen DEGs are common between lung carcinoma and other lung diseases associated with the recovery of tumoral characteristics. By using protein-protein interaction network we identified 10 hub genes including RHOA, CREB1, ACLY, AGO2, DNMT3A, CD44, BCL6, MMP3, PSMA6 and HNRNPA2B1. The DEG-TF network and DEG-miRNA interactions network analyses disclosed variety of TFs (CREBBP, SP1, JUN and HDAC1) and miRNAs (has-mir-20a-5p, has-mir-155-5p, has-mir-34a-5p and has-mir-16-5p) that regulate as a key transcriptional and post-transcriptional regulators on hub DEGs. Depend on the expression of hub-DEGs the multivariate survival probability curves displayed the significant differences between the low-risk and high-risk groups in the SurvExpress web-tool and database, which reveal effective prognostic power of hub-DEGs. The comparison of the coexpression networks of carcinoma and different respiratory organ diseases allowed the identification of common property patterns with DEGs and TFs related to big tumoral processes and sign pathways, that haven´t been studied through an experiment, validate their role within the early stages of carcinoma.
Conclusion: Our studies identified and validated common cell pathways and regulatory biomolecules (TFs and miRNAs) that may contribute to the link between lung carcinoma and other lung diseases. Thus our data-driven approaches can produce new insights into disease interaction mechanisms.
Keywords
Molecular signature, Molecular pathway, Lung cancer, Lung diseases.
Introduction
One of the most common malignant tumors, lung cancer affects millions of people worldwide. It is the leading global cause of cancer death in both men and women [1]. Small Cell Cancer (SCLC) and Non-Small Cell Cancer (NSCLC), which together account for 85% of all lung malignancies, are the two primary histological forms of the disease [2]. According to breast cancer (11.6%), prostate cancer (7.1%), and colorectal cancer (6.1%), lung cancer is the most often diagnosed cancer (11.6% of all cases) and the leading cause of cancer death (18.4% of all cancer-related fatalities) [3]. Lung cancer is the most prevalent condition in men and the one that results in the most cancer-related fatalities. The rise in death rates over the past few decades has been linked to delayed diagnosis, which restricts treatment options and highlights the critical lack of biomarkers for the creation of targeted therapies for the disease [4].
One of the most prevalent illnesses among children is asthma. Asthma's primary symptom is chronic lung inflammation, which manifests as airway hyper-reactivity, excessive mucus production, and breathing obstruction. Chronic inflammation has a significant role in the emergence of cancer [5]. As a result, asthmatic persistent inflammation may promote the growth of lung cancer. According to some earlier research, lung cancer risk and asthma are significantly related [6-23].
In terms of disease burden, COPD ranked fifth globally [24] and was the third most prevalent cause of death globally in 2010 [25]. COPD is a progressive, ultimately deadly decline in lung capacity. Up to 50% of smokers may experience a significant decline in quality of life due to COPD [26]. Exogenous and endogenous oxidative stress, inflammatory cytokine release, protease activity (owing to the protease: Anti-protease imbalance), and autoantibody production all contribute to lung injury in COPD [27]. Airway damage, air entrapment, and lung hyperinflation can all result from these. There is evidence that lung cancer and COPD share a same etiology, and smoking is the main contributor. COPD is a separate risk factor for lung cancer, specifically squamous cell carcinoma [28], and smokers with airflow restriction are up to five times more likely to develop lung cancer than those with adequate lung function [29]. The high incidence of lung cancer in COPD raises the possibility of shared processes, including early aging of the lungs, hereditary susceptibility to illness, or shared pathogenic agents, including growth factors, activation of intracellular pathways, or epigenetics [30].
The main airways of the lungs (bronchi) are affected by the infectious condition known as bronchitis. Most cases of bronchitis occur when an infection irritates and inflames the airways, causing them to generate more mucus than usual. After taking into account smoking and other respiratory conditions, chronic bronchitis and emphysema were found to be strongly related with lung cancer [31]. Instead of being an infrequent risk, chronic bronchitis may play a significant role in the development of lung cancer.
The SARS-CoV-2 virus is the infectious disease known as COVID-19. One of the most significant public health issues currently affecting our society is the recent COVID outbreak, which has caused more than 5,879,215 fatalities worldwide and incalculable economic harm. The World Health Organization classified COVID-19 to be a global pandemic on March 12, 2020, as the epidemic condition grew worse. In fact, a number of clinically significant prognostic factors, including advancing age, male gender, and smoking status, were linked to an increased 30- day all-cause mortality of COVID-19 (former smoker). Retrospective investigation proved that cancer patients in the outbreak city were 2.31 times more likely to contract a new coronavirus infection than the general population. Additionally, after COVID-19 infection, those with tumors or other problems had a worse prognosis than the general population [32].
Genomic studies provide the knowledge needed to identify all transcriptionally deregulated gene groupings. When comparing tumoral and healthy tissue, DEGs from microarrays and RNA-Seq have the capacity to participate in the control of biological activities and signaling networks, explaining expression patterns linked to tumor grade and patient survival [33-35]. According to our research team, a combined bioinformatics analysis of a particular subset of these studies can make use of all the transcriptomic knowledge produced by these technologies, uncovering potentially useful data that could be used to improve understanding of cellular processes connected to complex diseases like cancer. The deregulated genes were discovered in order to understand the intricacy of lung cancer on a worldwide scale. Additionally, when comparing various pathologies using a global analysis of numerous databases, we are able to identify particularities and shared processes that are impossible to identify when studying each study independently. In this study, we talk about how lung disease genetics affect lung cancer.
Materials and Methods
Overview of this study
This study evaluates the comorbidity interaction of Differentially Expressed Gene (DEGs) datasets using a quantitative systematic approach. In this strategy, gene expression analytics are combined with data on proteinprotein interactions, disease-gene connections, signal pathway information, Gene Ontology (GO) data, and gene expression analytics that are validated using these sources. In this research, the common elements to the comorbidities of putative pathways are found, along with the pathways' known pathogenic potential.
Microarray data collection and processing
We first examined gene expression microarray datasets to look at shared molecular routes to lung cancer with asthma, bronchitis, COPD, and COVID. Data on transcriptomes was gathered from the NCBI-GEO database, which is openly accessible [36]. Users can obtain the gene expression profiles kept in GEO using the datasets from numerous experiments that have been deposited in this database. Using specialized libraries from the R/Bioconductor language [37], six microarray datasets (GSE33532 and GSE99316 for lung cancer, and GSE143303 for asthma, GSE74163 for bronchitis, GSE133096 for COPD, and GSE163529 for COVID) were collected and compared to normal and disease-related lung tissue in order to find Differentiated Expressed Genes (DEGs) shared by lung pathologies and lung cancer as well. Then, the DEGs between the case and control samples were determined using the LIMMA statistical test [37]. The P-values were modified using Benjamini Hochberg's approach in order to regulate the multiple-testing false discovery rate [38]. The adjusted P-value and log2FC values were both taken into account when determining which DEGs were upregulated and downregulated. Upregulated DEGs, if adjusted P<0.001 and log2 FC>1 and downregulated DEGs, if adjusted P<0.001 and log2 FC<-1.
Enrichment analysis by DAVID
For the investigation of KEGG pathway enrichment and functional annotation of GO, we employed the web-based DAVID v6.8 program [39]. DAVID is a key resource for functional evaluation of high throughput gene expression patterns. In order to observe the DEGs involved in the GO keywords and pathways we effectively utilized, integrative analysis using the DAVID software. Analysis of the initial GEO2R DEGs revealed shared DEGs between lung cancer and other lung disorders. The study also analyzes molecular function, biological process and cellular component of GO enrichment analysis. This analysis was conducted by common DEGs, and P<0.05 were considered to be significant.
Protein-protein interaction network analysis of DEGs
The Protein-protein Interaction (PPI) network of the proteins expressed by DEGs was built using the STRING database [40]. A score combiner based on the product of probabilities is used by the STRING database [41]. Network analyst [42] was used to visualize and carry out topological analysis on the PPI network. Through the CytoHubba plugin [43] in Cytoscape 3.8.2, the topological analysis was used to identify hub-DEGs/proteins while taking both degree (connectivity) and betweenness metrics into account. In CytoHubba, a minimum degree of 10 was regarded as the cutoff threshold.
Mutation analysis
To evaluate the mutation or genomic alteration of hub DEGs, the online SRplot database (https://www. bioinformatics.com.cn/plot_basic_maf_summary_ plot_134_en ) was used. A TCGA-defined file format called Mutation Annotation Format (MAF) is used to contain information about mutation annotation. It provides SNV and annotation information for all samples, making it simple for downstream analysis. MAF is a particular file format for the human species. The output of this analysis displayed the most significant mutation or genomic alteration of hub DEGs.
DEG-TF interaction network analysis
We looked into the network of interactions between Transcription Factors (TFs), DEGs, and miRNAs in order to uncover the transcriptional regulatory components of these DEGs by using the publicly available miRNet database [44]. As the regulators of the discovered DEGs, the top TFs and miRNAs with the highest topological matrix degree were chosen.
Cross-validation and performance assessment of reported biomolecules
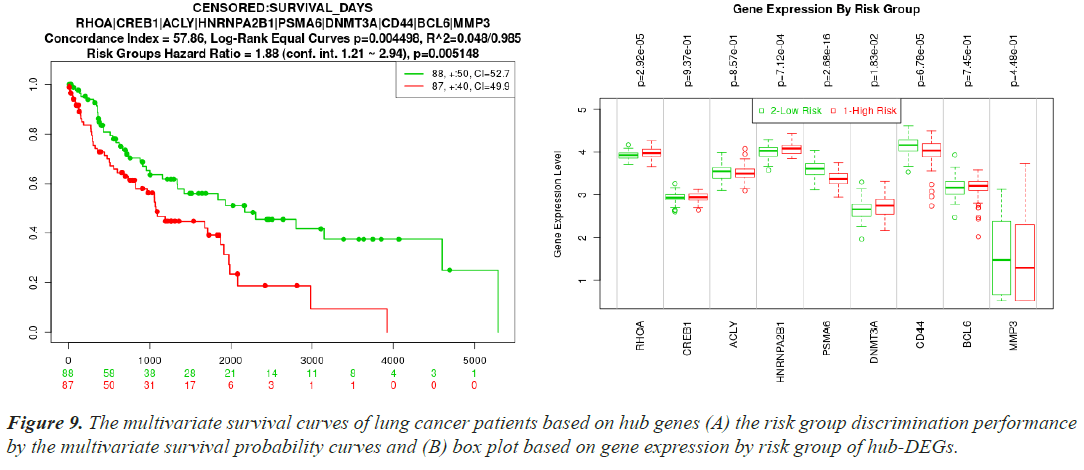
In the SurvExpress web server, patients were initially split into low-risk and high-risk groups [45]. Then, using box plots and survival probability curves, the differences between the risk groups based on the hub-DEG expression levels were examined. The t-test was used to assess the statistical significance of the differences in the box plots. Kaplan-Meier plots were used to assess the survival patterns of the reported biomolecules, and all survival studies required a log-rank P<0.01 to be considered statistically significant.
Results
Common DEGs between lung cancer and other lung diseases
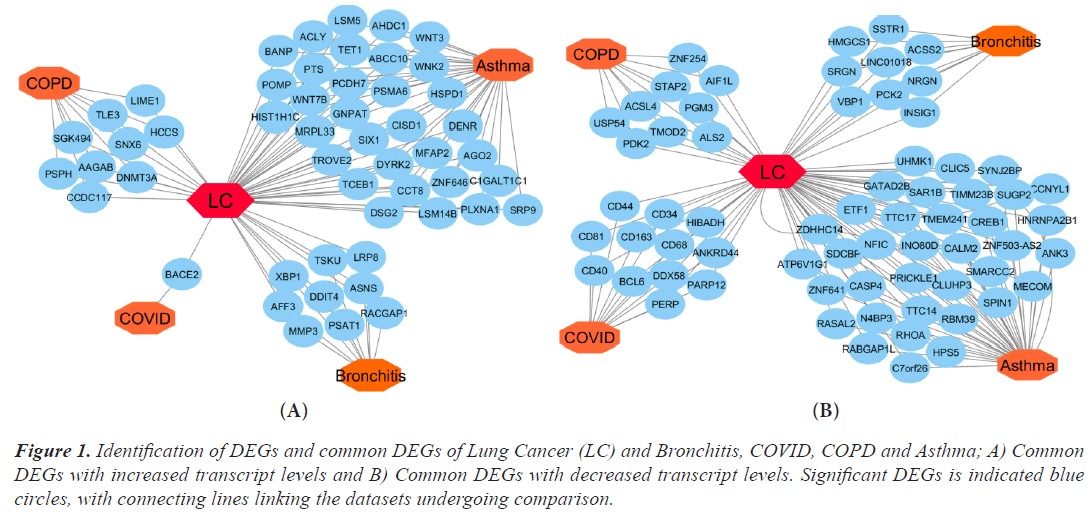
Using Limma, DEGs were examined in the microarray datasets and chosen from comparisons between the disease and control (Bioconductor packages). The R Bioconductor tools statistically assessed the DEGs that were discovered for each dataset. There were 356 DEGs found for lung cancer, 1035 for asthma, 287 for bronchitis, 1276 for COPD, and 34 for COVID datasets, respectively. In order to find the relevant genes that are shared by Lung Cancer (LC) and other lung illnesses, we also ran comparison analysis. Asthma, bronchitis, COPD, and COVID diseases, respectively, share 38, 9, 9, and 12 significantly down-regulated genes and 32, 9, 9, and 1 significantly up-regulated gene with LC. We built an upand down-regulation illness interaction network focused on lung cancer where four diseases are comorbid in order to find statistically significant relationships between lung cancer and the other ailments (Figure 1A and 1B).
Figure 1: Identification of DEGs and common DEGs of Lung Cancer (LC) and Bronchitis, COVID, COPD and Asthma; A) Common DEGs with increased transcript levels and B) Common DEGs with decreased transcript levels. Significant DEGs is indicated blue circles, with connecting lines linking the datasets undergoing comparison.
Common functional enrichment analysis in lung cancer and other lung diseases
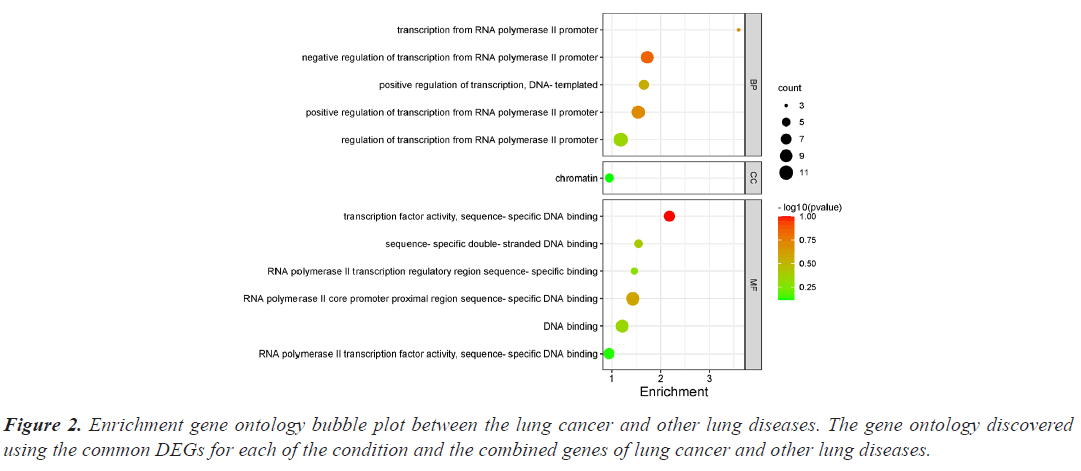
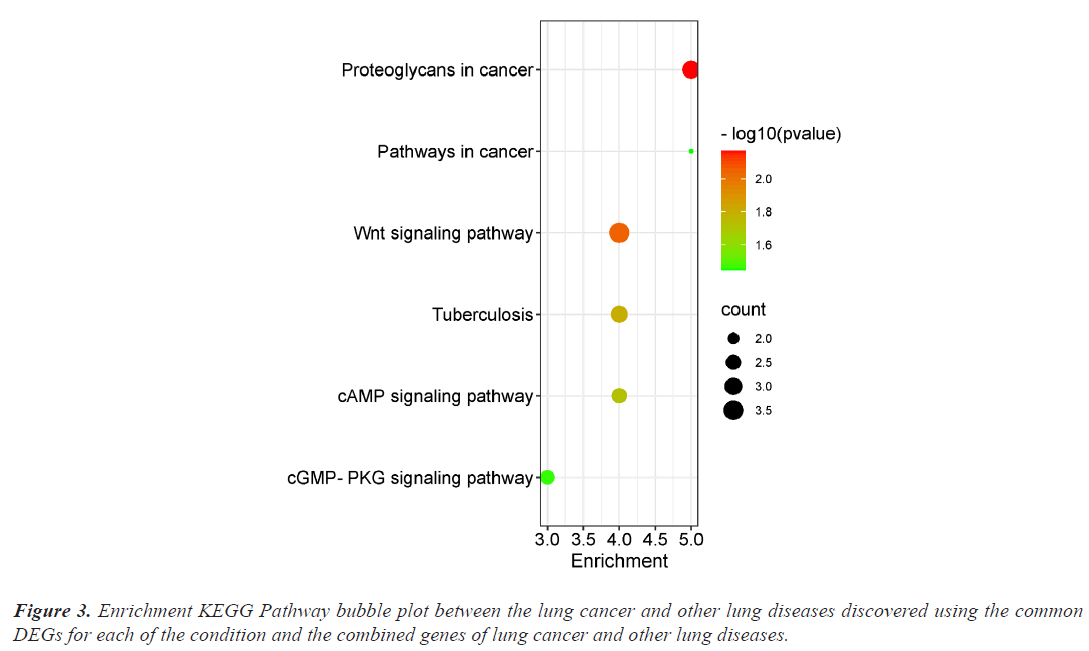
The investigation of the functional annotation of DEGs was conducted using the DAVID v6.8 web server. Approximating Biological Process (BP), Molecular Functions (MF), and Cellular Component (CC) terms were used to find the probable Gene Ontology (GO) classification. Signaling pathways are discussed in KEGG pathway enrichment terms. The adjusted P-value (adj P-value) ≤ 0.05 and FDR<0.05 are considered strongly enriched and result are presented in bubble plot (Figures 2 and 3). The Gene Ontology BP study demonstrated the important involvement of DEGs in transcription from RNA polymerase II promoter regulation of transcription, including positive regulation of transcription and negative regulation of transcription. We discovered using MF analysis that the DEGs from the complex network were enriched in RNA polymerase II transcription factor activity sequence-specific DNA binding, DNA binding, and RNA polymerase II core promoter proximal region sequencespecific DNA binding. In CC, the chromatin network was active. We used KEGG data enrichment for lung cancer and other lung illnesses to determine the pathways. After combining transcriptome and proteome studies, we carried out a regulation study to learn more about the pathways connected to these frequently occurring DEGs. The KEGG analysis showed that DEGs that are considerably enriched are involved in the Wnt signaling pathway, Proteoglycans in cancer, Pathways in cancer, Tuberculosis, and cAMP signaling pathway.
Identification of hub DEGs of lung cancer in common with other lung diseases
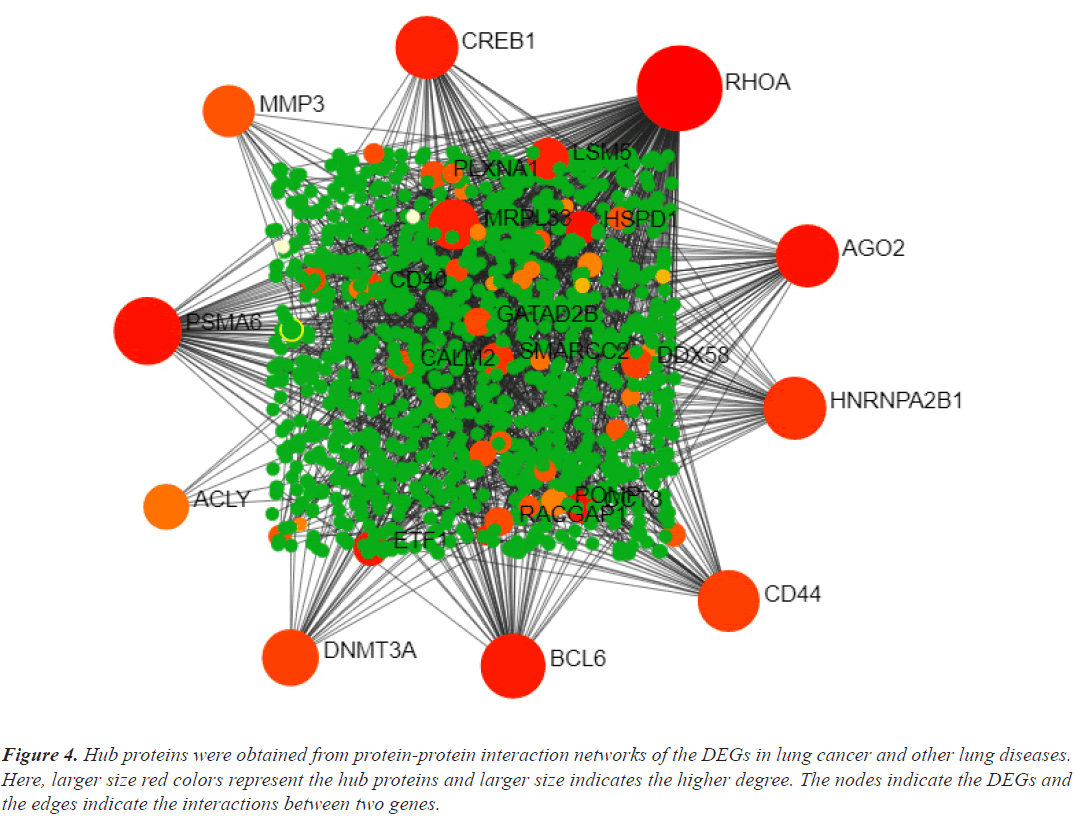
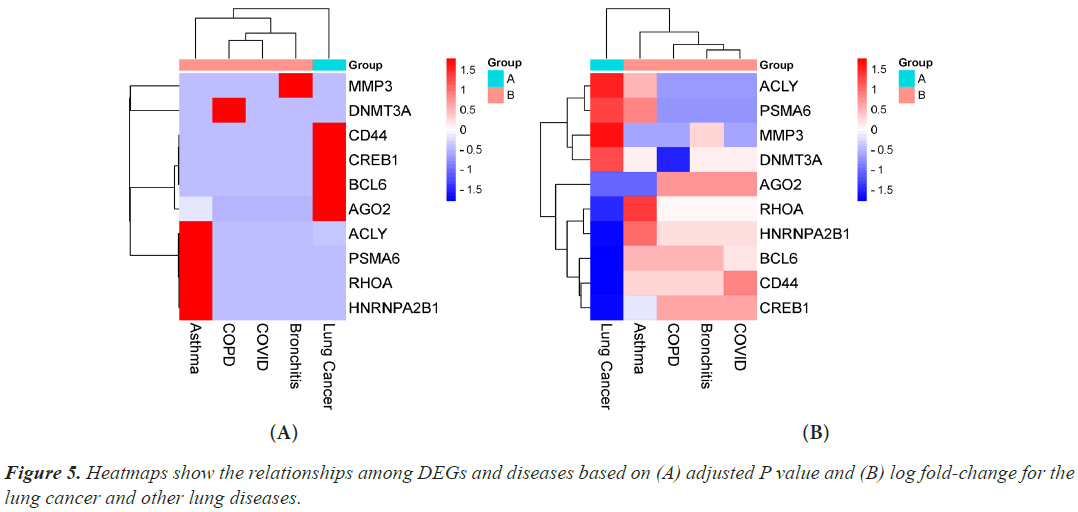
Hub genes are important for signal transduction activities that occur as a disease progresses. Through physical interactions and topological analysis, a PPI subnetwork was built around the proteins expressed by the DEGs in order to identify these hub genes. The ten hub genes identified as a result are RHOA, CREB1, ACLY, HNRNPA2B1, AGO2, PSMA6, DNMT3A, CD44, BCL6 and MMP3 (Figure 4). Using rank of degree the connection between hub genes and nodes was calculated and red color nodes are highly connected. RHOA has the highest degree score (218), followed by PSMA6, BCL6, AGO2, HNRNPA2, and CREB1 (Table 1). Figure 5A heat map illustrated the association between hub genes and diseases in terms of the adj P-value, whereas Figure 5B heat map illustrates the association between hub genes and diseases in terms of the values of log fold change in the SRplot database. By analysis this study, lung cancer dataset shares more DEGs with asthma compared to the other three diseases (bronchitis, COPD and COVID). The DEGs that are common between lung cancer and asthma include RHOA, CREB1, ACLY, HNRNPA2, AGO2 and PSMA6 (Figure 6). The MMP3 shared with bronchitis and lung cancer and DNMT3A is a common DEG between COPD and lung cancer (Figure 6). Two DEGs share with COVID and lung cancer include CD44 and BCL6 (Figure 6).
| Rank | Gene name | Score | Log FC | Adjusted P-value |
|---|---|---|---|---|
| 1 | RHOA | 218 | -0.4477457 | 4.59E-09 |
| 2 | PSMA6 | 66 | 0.6102423 | 7.42E-07 |
| 3 | BCL6 | 52 | -1.2212194 | 2.35E-23 |
| 4 | AGO2 | 44 | 1.1021344 | 3.00E-06 |
| 4 | HNRNPA2B1 | 44 | 0.179547 | 1.21E-02 |
| 4 | CREB1 | 44 | 0.2207258 | 1.52E-02 |
| 5 | CD44 | 40 | -0.43077 | 2.27E-02 |
| 6 | DNMT3A | 25 | 0.9477237 | 5.13E-13 |
| 7 | MMP3 | 15 | 2.11972 | 2.64E-06 |
| 8 | ACLY | 6 | 0.446152 | 2.81E-05 |
Table 1. Top ten hub genes network from protein-protein interaction string database ranked by degree method.
Figure 4: Hub proteins were obtained from protein-protein interaction networks of the DEGs in lung cancer and other lung diseases. Here, larger size red colors represent the hub proteins and larger size indicates the higher degree. The nodes indicate the DEGs and the edges indicate the interactions between two genes.
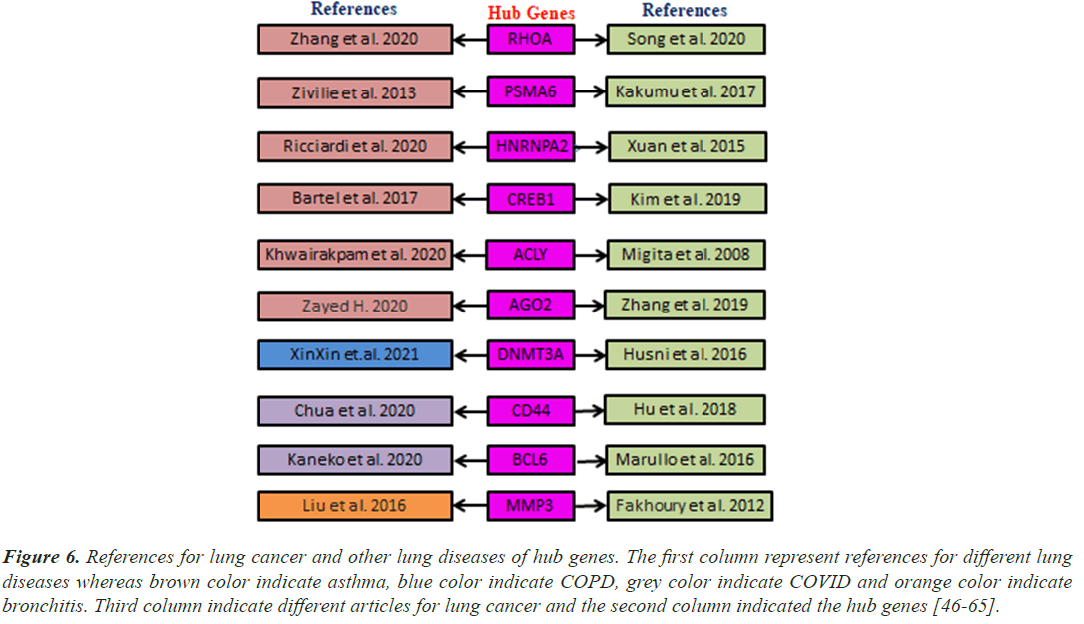
Figure 6: References for lung cancer and other lung diseases of hub genes. The first column represent references for different lung diseases whereas brown color indicate asthma, blue color indicate COPD, grey color indicate COVID and orange color indicate bronchitis. Third column indicate different articles for lung cancer and the second column indicated the hub genes [46-65].
Mutation analysis of hub genes
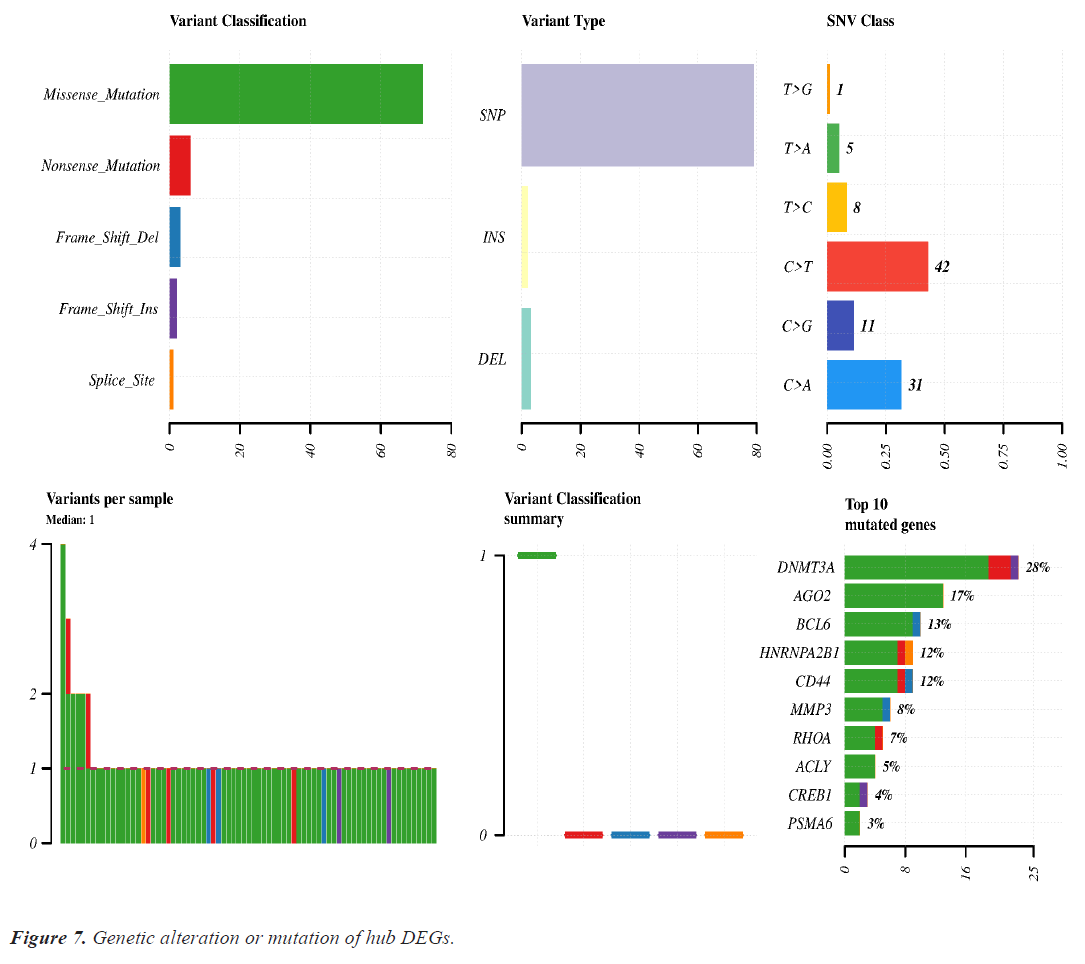
Mutation analysis or genomic alteration of 10 hub genes disclose that DNMT3A, AGO2, BCL6, HARNPA2B1, CD44 and MMP3 genes had 28%, 17%, 13%, 12%, 12% and 8% mutation over the lung cancer studies. Other hub genes had low rate mutation in lung cancer. Maximum genomic alteration are happened due to missense mutation. Details of the genomic alteration of hub genes are present in Figure 7.
Transcriptional and post-transcriptional regulators analysis
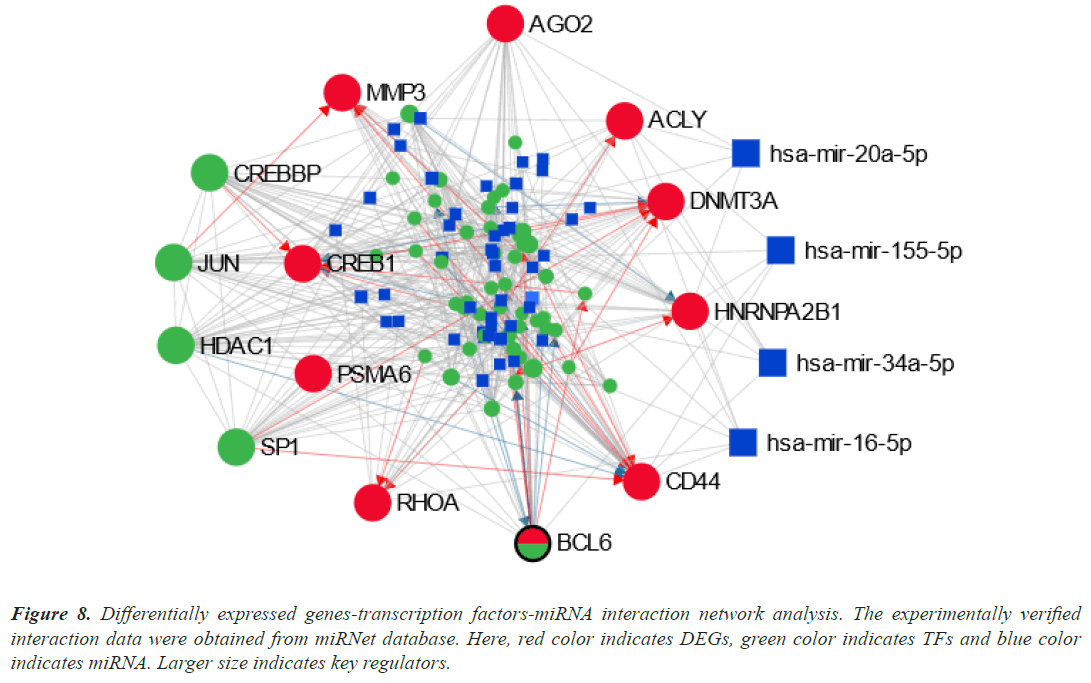
We examined the TFs-DEGs and miRNAs-DEGs networks in miRNet to find the DEGs' transcriptional and posttranscriptional regulators (Figure 8). The TFs (CREBBP, SP1, JUN, and HDAC1) and miRNAs (hasmir- 20a-5p, has-mir-155-5p, has-mir-34a-5p, and has-mir- 16-5p) were shown to be the key regulators of the DEGs that were identified as shared by lung cancer and other lung disorders by statistical analysis of the topological parameters.
Figure 8: Differentially expressed genes-transcription factors-miRNA interaction network analysis. The experimentally verified interaction data were obtained from miRNet database. Here, red color indicates DEGs, green color indicates TFs and blue color indicates miRNA. Larger size indicates key regulators.
Cross-validation and risk discrimination performance of hub proteins
A separate RNA-seq dataset collected from TCGA was used to confirm the differential expression signatures, and the effectiveness of 10 hub proteins in risk discrimination was assesseds. The risk discrimination performance and the differential expression pattern were noticed by the online gene validation website SurvExpress. Depending on how well they could identify between low-risk and high-risk situations, the samples were split into two groups based on expression levels, and these groups were given the names low-risk and high-risk. Nine genes were accessible in the TCGA Lung Squamous Cell Carcinoma Survival Information Database following the hub gene upload, but AGO2 was not. The box plot of their gene expressions and the survival curves for the high-risk and low-risk groups are shown in (Figure 9A and 9B). For both investigations, the prognostic index, log-rank test, and hazard ratio are shown.
Discussion
Our investigation will contribute to a better understanding of the potential influences that other lung conditions may have on the emergence of lung cancer. Instead of taking a purely mechanistic approach, we used a bioinformatics tool to hunt for genes that were dysregulated in lung cancer and other lung disorders. We then used this information to provide hints to find dysregulated pathways and control mechanisms. In this study, we examine the relationship between four lung diseases: Asthma, bronchitis, COPD, and COVID-19 and lung cancer. The analysis's potency and quality can also be enhanced over time as new, sizable gene expression datasets related to diseases become available. The significant biomarker profiles have been linked to a higher risk of getting cancer [66]. Our disease network identified a number of shared genes between lung cancer and other diseases using a combined study of transcriptome, genomic, PPI, pathway, and GO data.
This study focuses on identifying potential paths of communication between lung cancer and other lung disorders. First, we determined which genes from each dataset had differential expression. Then, we contrasted the DEGs of the datasets for lung cancer and the four other lung conditions. There were 356 DEGs found for lung cancer, 1035 for asthma, 287 for bronchitis, 1276 for COPD, and 34 for COVID datasets respectively. In order to find the relevant genes that are shared by Lung Cancer (LC) and other lung illnesses, we also ran comparison analysis. Asthma, bronchitis, COPD, and COVID diseases, respectively, share 38, 9, 9, and 12 significantly down-regulated genes and 32, 9, 9, and 1 significantly upregulated genes with LC (Figure 1). Functional enrichment are involved in some cancer related pathways including Wnt signaling pathway, Proteoglycans in cancer, Pathways in cancer, Tuberculosis, and cAMP signaling pathway (Figure 3).
Caucasians and Asians both have a much higher risk of lung cancer when they have asthma, and both males and female patients exhibit this increased risk [67]. Although smoking is known to raise the risk of lung cancer, asthma patients who do not smoke also have an elevated risk of lung cancer [68]. These findings suggested that lung cancer and asthma may both be at risk. Lung cancer may have long-term lung inflammation caused by asthma as a contributing factor. We discovered 70 DEGs between lung cancer and asthma in this study. Next, we found ten hub genes (Figure 4), of which six DEGs (RHOA, CREB1, ACLY, HNRNPA2, AGO2, and PSMA6) had a substantial association with both lung cancer and asthma (Figure 5). After an initial but temporary dysregulation of CREB1 and CRTC-mediated transcription is either directly or indirectly acceptable to influence long-term modifications of downstream targets in asthma, down-regulated CREB1 was profoundly affected in early primary human bronchial epithelial cells [69]. Effective biomarkers for the diagnosis and prognosis of non-smokers with lung cancer include CREB1 [70]. According to the MTS tetrazolium assay, flow cytometry analysis, and western blotting, the inhibition of RhoA expression significantly reduced the proliferation of lung cancer cells while significantly increasing apoptosis (P<0.01). Additionally, the knockdown of RhoA results in significantly lower levels of phosphorylated signal transducer and activator of transcription (phospho-STAT3; P<0.01) and significantly higher levels of caspase-3 (P<0.01) in the cells [71]. Patients with asthma have higher levels of RhoA/Rhokinase activation, which is strongly correlated with airway hyper responsiveness and other important characteristics of asthma, such as airway remodeling, allergic airway inflammation, and malfunction of the airway barrier [72]. Human lung cancer samples were found to have much greater levels of phosphorylated ACLY overexpression than did normal lung tissue [73]. AGO2 deletion slows the growth of tumors, reduces their pathologic severity, and blocks KRAS signaling. Future therapeutic advancements in treating NSCLC and other KRAS-driven cancers may involve focusing on the AGO2-KRAS interaction. Comparing a panel of normal and lung cancer cell lines' putative proteasome subunit genes' expression analysis and gene copy number analysis, PSMA6 copy number expressed considerably higher levels of PSMA6 mRNA [74].
Lung cancer risk may be independently increased by COPD. Lung cancer may be caused by lung oxidative stress, persistent exposure to pro-inflammatory cytokines, increased cellular proliferation, and suppression of DNA repair processes [75]. When it comes to lung adenocarcinoma, DNMT3A expression is a standalone prognostic indicator that is connected with the lepidic subtype and histologically non-invasive type. The clinical use of DNMT3A as a significant prognostic predictor is possible. This suggests that DNMT3A is a rate-limiting factor preventing inflammatory reactions since DNMT3A expressing T helper cells might employ the same method to prevent immunological overreaction.
Lung cancer and bronchitis share many of the same risk factors, genetic and epigenetic abnormalities, activation of related signaling pathways, and poor prognoses. Lung cancers have much higher MMP3 levels than nonmalignant lung tissue [76]. The association between one location of the 6A-G haplotype analyzed by MMP3 (1171 5A>6A) and chronic bronchitis [77].
Patients with lung cancer and hematological malignancies appear to be at the highest risk of dying from SARSCoV- 2 infection. Lung cancer offers a clinical setting that is characterized by an increased risk of pulmonary complications, severe lung injury, and substantial COVID-19 mortality because of pathophysiological, clinical, and treatment-related risk factors. BCL6 was elevated in KRAS-mutant cancers, such as Non-Small- Cell Lung Cancer (NSCLC), following BET inhibition [78]. Lack of germinal center formation and Bcl-6 expression, which are both accompanied by elevated TNF-alpha production in secondary lymphoid organs, may compromise the humeral response to SARS-CoV-2 [63]. High levels of CD44 expression may hasten the development of NSCLC by boosting the proliferation of cancer cells [79]. Ten hub genes validated by using previous bibliography that these genes are responsible for lung cancer and other lung diseases (Figure 6).
Genomic alteration or mutation of hub DEGs shows that DNMT3A and AGO2 were mutated more than the other DEGs. Missense mutation is significantly associated with all genes mutation (Figure 7).
Our results identified four TFs (CREBBP, SP1, JUN, and HDAC1) and four miRNAs (has-mir-20a-5p, has-mir- 155-5p, has-mir-34a-5p, and has-mir-16-5p) as a key biomolecule regulators (Figure 8). CREBBP depletion in SCLC decreases histone acetylation and cellular adhesion gene transcription while promoting carcinogenesis. In the absence of c-Jun, the protein ranges of the JunD family member have increased. JunD phosphorylation increased in c-Jun-deficient cells, and introduction of a dominantenergetic JNKK2-JNK1 transgene also increased lung tumor development [80]. In vivo lung adenocarcinoma cell migration, invasion, and metastasis were all negatively impacted by Sp1. Due to Sp1's inherent instability, low Sp1 levels were also seen in highly invasive cancer cells [81]. One important epigenetic component that has been connected to the development and prognosis of various cancers is Histone Deacetylase 1 (HDAC1). HDAC1 expression was strongly correlated with the lung cancer differentiation grade, suggesting that it may be an important factor in this process [82].
Using multivariate survival probability curves and boxplots, the reported biomolecules' prognostic ability to distinguish between high-risk and low-risk scenarios were demonstrated (Figure 9). The described biomolecules certainly played a significant effect in patient survival, as seen by the survival curves. The molecular candidate's gene expression data box plot also clearly distinguished between the high-risk and low-risk groups.
Conclusion
Our research has uncovered and verified similar cell pathways and regulatory biomolecules that could play a role in the association between lung cancer and other lung conditions. RHOA, CREB1, ACLY, AGO2, DNMT3A, CD44, BCL6, MMP3, PSMA6, and HNRNPA2B1 are just a few of the strong candidate genes in common pathways that are linked to the recovery of tumoral features in lung cancer and other lung disorders. Numerous TFs (CREBBP, SP1, JUN, and HDAC1) and miRNAs (has-mir-20a-5p, has-mir-155-5p, has-mir-34a-5p and has-mir-16-5p) were identified through the analysis of the DEG-TF network and DEG-miRNA interactions network. As a result, the disease interaction processes can be better understood due to our data-driven methodologies. Lung diseases have a significant genetic influence on the lung cancer.
Author’s Contributions
Md. Nurul Haque Mollah: Conceptualization, Methodology, Supervision. S. M. Shahinul Islam: Supervision. Amina Rownaq: Methodology, Formal analysis, Writing-Original draft preparation.
Conflicts of Interest
There is no existing conflict of interest. We want to make sure there are no existing conflicts of interest related to this publication.
Acknowledgement
This research facility was made possible by the Institute of Biological Sciences at the University of Rajshahi in Rajshahi, Bangladesh, and the Lab of Bioinformatics at the Department of Statistics at the same university.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin 2015; 65: 5-29.
[Crossref] [Google Scholar] [PubMed]
- Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R. Non-small-cell lung cancer. Nat Rev Dis Primers 2015; 1: 15009.
[Crossref] [Google Scholar] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394-424.
[Crossref] [Google Scholar] [PubMed]
- Dela Cruz CS, Tanoue LT, Matthay RA. Lung cancer: Epidemiology, etiology, and prevention. Clin Chest Med 2011; 32: 605-644.
[Crossref] [Google Scholar] [PubMed]
- Qu YL, Liu J, Zhang LX, Wu CM, Chu AJ, Wen BL, Ma C, Yan XY, Zhang X, Wang DM, Lv X, Hou SJ. Asthma and the risk of lung cancer: A meta-analysis. Oncotarget 2017; 8: 11614-11620.
[Crossref] [Google Scholar] [PubMed]
- Alderson M. Mortality from malignant disease in patients with asthma. Lancet 1974; 2: 1475-1477.
[Crossref] [Google Scholar] [PubMed]
- Reynolds P, Kaplan GA. Asthma and cancer. Am J Epidemiol 1987; 125: 539-540.
[Crossref] [Google Scholar] [PubMed]
- Mills PK, Beeson WL, Fraser GE, Phillips RL. Allergy and cancer: Organ site-specific results from the adventist health study. Am J Epidemiol 1992; 136: 287-295.
[Crossref] [Google Scholar] [PubMed]
- Vesterinen E, Pukkala E, Timonen T, Aromaa A. Cancer incidence among 78,000 asthmatic patients. Int J Epidemiol 1993; 22: 976-982.
[Crossref] [Google Scholar] [PubMed]
- Eriksson NE, Holmen A, Hogstedt B, Mikoczy Z, Hagmar L. A prospective study of cancer incidence in a cohort examined for allergy. Allergy 1995; 50: 718-722.
[Crossref] [Google Scholar] [PubMed]
- Huovinen E, Kaprio J, Vesterinen E, Koskenvuo M. Mortality of adults with asthma: A prospective cohort study. Thorax 1997; 52: 49-54.
[Crossref] [Google Scholar] [PubMed]
- Boffetta P, Ye W, Boman G, Nyre n. Lung cancer risk in a population-based cohort of patients hospitalized for asthma in Sweden. Eur Respir J 2002; 19: 127-133.
[Crossref] [Google Scholar] [PubMed]
- Talbot-Smith A, Fritschi L, Divitini ML, Mallon DF, Knuiman MW. Allergy, atopy, and cancer: A prospective study of the 1981 Busselton cohort. Am J Epidemiol 2003; 157: 606-612.
[Crossref] [Google Scholar] [PubMed]
- vandentorren S, Baldi I, Annesi Maesano I, Charpin D, Neukirch F, Filleul L, Cantagrel A, Tessier JF. Long-term mortality among adults with or without asthma in the PAARC study. Eur Respir J 2003; 21: 462-467.
[Crossref] [Google Scholar] [PubMed]
- Littman AJ, Thornquist MD, White E, Jackson LA, Goodman GE, Vaughan TL. Prior lung disease and risk of lung cancer in a large prospective study. Cancer Causes Control 2004; 15: 819-827.
[Crossref] [Google Scholar] [PubMed]
- Turner MC, Chen Y, Krewski D, Ghadirian P, Thun MJ, Calle EE. Cancer mortality among US men and women with asthma and hay fever. Am J Epidemiol 2005; 162: 212-221.
[Crossref] [Google Scholar] [PubMed]
- Brown DW, Young KE, Anda RF, Felitti VJ, Giles WH. Re: Asthma and the risk of lung cancer. findings from the Adverse Childhood Experiences (ACE). Cancer Causes Control 2006; 17: 349-350.
[Crossref] [Google Scholar] [PubMed]
- González-Pérez A, Fernández-Vidaurre C, Rueda A, Rivero E, García Rodríguez LA. Cancer incidence in a general population of asthma patients. Pharmacoepidemiol Drug Saf 2006; 15: 131-138.
[Crossref] [Google Scholar] [PubMed]
- Ji J, Shu X, Li X, Sundquist K, Sundquist J, Hemminki K. Cancer risk in hospitalised asthma patients. Br J Cancer 2009; 100: 829-833.
[Crossref] [Google Scholar] [PubMed]
- Çolak Y, Afzal S, Nordestgaard BG, Lange P. Characteristics and prognosis of never-smokers and smokers with asthma in the copenhagen general population study. A Prospective Cohort Study. Am J Respir Crit Care Med 2015; 192: 172-181.
[Crossref] [Google Scholar] [PubMed]
- Huang JY, Jian ZH, Nfor ON, Ku WY, Ko PC, Lung CC, Ho CC, Pan HH, Huang CY, Liang YC, Liaw YP. The effects of pulmonary diseases on histologic types of lung cancer in both sexes: A population-based study in Taiwan. BMC Cancer 2015; 15: 834.
[Crossref] [Google Scholar] [PubMed]
- Fan Y, Jiang Y, Hu P, Chang R, Yao S, Wang B, Li X, Zhou Q, Qiao Y. Modification of association between prior lung disease and lung cancer by inhaled arsenic: A prospective occupational-based cohort study in Yunnan, China. J Expo Sci Environ Epidemiol 2016; 26: 464-470.
[Crossref] [Google Scholar] [PubMed]
- Pirie K, Peto R, Green J, Reeves GK, Beral V. Lung cancer in never smokers in the UK million women study. Int J Cancer 2016; 139: 347-354.
[Crossref] [Google Scholar] [PubMed]
- Vestbo J, Hurd SS, Alvar GA. Global Strategy for the diagnosis, management and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013; 187: 347-365.
[Crossref] [Google Scholar] [PubMed]
- Burney PGJ, Patel J, Newson R. Global and regional trends in COPD mortality, 1990-2010. Eur Respir J 2015; 45: 1239-1247.
[Crossref] [Google Scholar] [PubMed]
- Lundbäck B, Lindberg A, Lindstrom M. Not 15 but 50% of smokers develop COPD? report from the obstructive lung disease in northern sweden studies. Respir Med 2003; 97: 115-122.
[Crossref] [Google Scholar] [PubMed]
- Brusselle GG, Joos GF, Bracke KR. New insights into the immunology of chronic obstructive pulmonary disease. Lancet 2015; 378: 1015-1026.
[Crossref] [Google Scholar] [PubMed]
- Papi A. COPD increases the risk of squamous histological subtype in smokers who develop non-small cell lung carcinoma. Thorax 2004; 59: 679-681.
[Crossref] [Google Scholar] [PubMed]
- Young RP, Hopkins RJ. Link between COPD and lung cancer. Respir Med 2010; 104: 758-759.
[Crossref] [Google Scholar] [PubMed]
- Barnes PJ, Adcock IM. Chronic obstructive pulmonary disease and lung cancer: A lethal association. Am J Respir Crit Care Med 2011; 184: 866-867.
[Crossref] [Google Scholar] [PubMed]
- Denholm R, Schüz J, Straif K, Stücker I. Is previous respiratory disease a risk factor for lung cancer? Am J Respir Crit Care Med 2014; 190: 549-559.
[Crossref] [Google Scholar] [PubMed]
- Liang W, Guan W, Chen R. Cancer patients in SARS-CoV-2 infection: A nationwide analysis in China. Lancet Oncol 2020; 21: 335-337.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Liotta L. Clinical bioinformatics: A new emerging science. J Clin Bioinforma 2011; 1: 1.
[Crossref] [Google Scholar] [PubMed]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603-607.
[Crossref] [Google Scholar] [PubMed]
- Ma X, Zhang S, Shang Q, Long S, Piao X. Determination and prediction of the apparent and standardized ileal amino acid digestibility in cottonseed meals fed to growing pigs. Anim Sci J 2019; 90: 655-666.
[Crossref] [Google Scholar] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207-210.
[Crossref] [Google Scholar] [PubMed]
- Durinck S, Spellman P, Birney E. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009; 4: 1184-1191.
[Crossref] [Google Scholar] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to Multiple Testing. J R Stat Soc Series Stat Methodol 1995; 57: 289-300.
[Crossref] [Google Scholar] [PubMed]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 2009; 4: 44-57.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017; 45: D362–D368.
[Crossref] [Google Scholar] [PubMed]
- Li X, Li W, Zeng M, Zheng R, Li M. Network-based methods for predicting essential genes or proteins: A survey. Brief Bioinform 2020; 21: 566-583.
[Crossref] [Google Scholar] [PubMed]
- Xia J, Gill EE, Hancock REW. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015; 10: 823-844.
[Crossref] [Google Scholar] [PubMed]
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014; 8: S11.
[Crossref] [Google Scholar] [PubMed]
- Chang L, Xia J. microRNA regulatory network analysis using miRNet 2.0. Methods Mol Biol 2023; 2594: 185-204.
[Crossref] [Google Scholar] [PubMed]
- Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, Martínez-Torteya A, Chacolla-Huaringa, R. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013; 8: e74250.
[Crossref] [Google Scholar] [PubMed]
- Song F, Xuan Z, Yang X, Ye X, Pan Z, Fang Q. Identification of key microRNAs and hub genes in non-small-cell lung cancer using integrative bioinformatics and functional analyses. J Cell Biochem 2020; 121: 2690-2703.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Saradna A, Ratan R, Ke X, Tu W, Do DC, Hu C, Gao P. RhoA/Rho-kinases in asthma: from pathogenesis to therapeutic targets. Clin Transl Immunology 2020; 9: e01134.
[Crossref] [Google Scholar] [PubMed]
- Kakumu T, Sato M, Goto D, Kato T, Yogo N, Hase T, Morise M, Fukui T. Identification of proteasomal catalytic subunit PSMA6 as a therapeutic target for lung cancer. Cancer Sci 2017; 108: 732-743.
[Crossref] [Google Scholar] [PubMed]
- Zivile Z, Edita G, Astra V, Brigita S, Raimundas S. Single nucleotide polymorphism of PSMA6 gene in Lithuanian patients with asthma. Eur Resp J 2013; 42: P1395.
- Xuan Y, Wang J, Ban L, Lu JJ, Yi C, Li Z. hnRNPA2/B1 activates cyclooxygenase-2 and promotes tumor growth in human lung cancers. Mol Oncol 2016; 10: 610-624.
[Crossref] [Google Scholar] [PubMed]
- Ricciardi L, Giurato G, Memoli D, Pietrafesa M, Dal Col J. Posttranscriptional gene regulatory networks in chronic airway inflammatory diseases: In silico mapping of rna-binding protein expression in airway epithelium. Front Immunol 2020; 11: 579889.
[Crossref] [Google Scholar] [PubMed]
- Kim IK, McCutcheon JN, Rao G, Liu SV, Pommier Y. Acquired SETD2 mutation and impaired CREB1 activation confer cisplatin resistance in metastatic non-small cell lung cancer. Oncogene 2019; 38: 180-193.
[Crossref] [Google Scholar] [PubMed]
- Bartel S, Schulz N, Alessandrini F, Schamberger AC, Pagel P. Pulmonary microRNA profiles identify involvement of Creb1 and Sec14l3 in bronchial epithelial changes in allergic asthma. Sci Rep 2017; 7: 46026.
[Crossref] [Google Scholar] [PubMed]
- Migita T, Narita T, Nomura K, Miyagi E, Inazuka F. ATP citrate lyase: Activation and therapeutic implications in non-small cell lung cancer. Cancer Res 2008; 68: 8547-8554.
[Crossref] [Google Scholar] [PubMed]
- Khwairakpam AD, Banik K, Girisa S, Shabnam B, Shakibaei M, Fan L. The vital role of ATP citrate lyase in chronic diseases. J Mol Med (Berl) 2020; 98: 71-95.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Wang Y, Dou J. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene 2019; 38: 1410-1431.
[Crossref] [Google Scholar] [PubMed]
- Zayed H. Novel comprehensive bioinformatics approaches to determine the molecular genetic susceptibility profile of moderate and severe asthma. Int J Mol Sci 2020; 21: 4022.
[Crossref] [Google Scholar] [PubMed]
- Husni RE, Shiba-Ishii A, Iiyama S, Shiozawa T, Kim Y, Nakagawa T, Sato T, Kano J, Minami Y, Noguchi M. DNMT3A expression pattern and its prognostic value in lung adenocarcinoma. Lung Cancer 2016; 97: 59-65.
[Crossref] [Google Scholar] [PubMed]
- XinXin Huo, SiHui Jin, YiGe Wang, Li Ma. DNA methylation in chronic obstructive pulmonary disease. Epigenomics 2021; 13: 1145-1155.
[Crossref] [Google Scholar] [PubMed]
- Hu B, Ma Y, Yang Y, Zhang L, Han H, Chen J. CD44 promotes cell proliferation in non-small cell lung cancer. Oncol Lett 2018; 15: 5627-5633.
[Crossref] [Google Scholar] [PubMed]
- Chua RL, Lukassen S, Trump S. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat Biotechnol 2020; 38: 970-979.
[Crossref] [Google Scholar] [PubMed]
- Marullo Rossella, Ahn Haelee, Cardenas Mariano. Abstract 1271: The transcription factor BCL6 is a rational target in Non-Small Cell Lung Cancer (NSCLC). Cancer Res 2016; 76: 1271.
- Kaneko N, Kuo HH, Boucau J, Farmer JR, Allard-Chamard H. Loss of Bcl-6-expressing T follicular helper cells and germinal centers in COVID-19. Cell 2020; 183: 143-157.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Yue Z, Yu J. Proteomic characterization reveals that MMP-3 correlates with bronchiolitis obliterans syndrome following allogeneic hematopoietic cell and lung transplantation. Am J Transplant 2016; 16: 2342-2351.
[Crossref] [Google Scholar] [PubMed]
- Fakhoury HM, Noureddine S, Tamim H, Chmaisse H, Makki R. Association of MMP3-1171(5A>6A) polymorphism with lung cancer in lebanon. Genet Test Mol Biomarkers 2012; 16: 988-990.
[Crossref] [Google Scholar] [PubMed]
- Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: Current insights and future perspectives. Nat Rev Cancer 2017; 17: 692-704.
[Crossref] [Google Scholar] [PubMed]
- Qu YL, Liu J, Zhang LX, Wu CM, Chu AJ, Wen BL, Ma C, Yan XY, Zhang X, Wang DM, Lv X, Hou SJ. Asthma and the risk of lung cancer: A meta-analysis. Oncotarget 2017; 8: 11614-11620.
[Crossref] [Google Scholar] [PubMed]
- Pirie K, Peto R, Green J. Lung cancer in never-smokers in the UK million women study. Int J Cancer 2016; 139: 347-354.
[Crossref] [Google Scholar] [PubMed]
- Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzás EI, Buck AH, de Candia P, Chow FW, Das S. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA-an ISEV position paper. J Extracell Vesicles 2017; 6: 1286095.
[Crossref] [Google Scholar] [PubMed]
- Zhang X, Gao C, Liu L, Zhou C, Liu C, Li J, Zhuang J, Sun C. DNA methylation-based diagnostic and prognostic biomarkers of nonsmoking lung adenocarcinoma patients. J Cell Biochem 2019; 120: 13520-13530.
[Crossref] [Google Scholar] [PubMed]
- Liu D, Mei X, Wang L, Yang X. RhoA inhibits apoptosis and increases proliferation of cultured SPCA1 lung cancer cells. Mol Med Rep 2017; 15: 3963-3968.
[Crossref] [Google Scholar] [PubMed]
- Chiba Y, Misawa M. The role of RhoA-mediated Ca2+ sensitization of bronchial smooth muscle contraction in airway hyperresponsiveness. J Smooth Muscle Res 2004; 40: 155-167.
[Crossref] [Google Scholar] [PubMed]
- Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, Ushijima M, Mashima T, Seimiya H, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y. ATP citrate lyase: Activation and therapeutic implications in non-small cell lung cancer. Cancer Res 2008; 68: 8547-8554.
[Crossref] [Google Scholar] [PubMed]
- Kakumu T, Sato M, Goto D, Kato T, Yogo N, Hase T, Morise M, Fukui T, Yokoi K. Identification of proteasomal catalytic subunit PSMA6 as a therapeutic target for lung cancer. Cancer Sci 2017; 108: 732-743.
[Crossref] [Google Scholar] [PubMed]
- Durham AL, Adcock IM. The relationship between COPD and lung cancer. Lung Cancer 2015; 90: 121-127.
[Crossref] [Google Scholar] [PubMed]
- Neha Merchant, Ganji Purnachandra Nagaraju, Balney Rajitha, Saipriya Lammata, Kishore Kumar Jella. Matrix metalloproteinases: Their functional role in lung cancer. Carcinogenesis 2017; 38: 766-780.
[Crossref] [Google Scholar] [PubMed]
- Korytina G, Akhmadishina L, Viktorova E. Extracellular matrix remodeling genes polymorphisms and risk of chronic bronchitis and recurrent pneumonia in children. J Hum Genet 2013; 58: 467-474.
[Crossref] [Google Scholar] [PubMed]
- Guo J, Liu Y, Lv J, Zou B, Chen Z, Li K, Feng J, Cai Z, Wei L, Liu M, Pang X. BCL6 confers KRAS-mutant non-small-cell lung cancer resistance to BET inhibitors. J Clin Invest 2021; 131: e133090.
[Crossref] [Google Scholar] [PubMed]
- Hu B, Ma Y, Yang Y, Zhang L, Han H, Chen J. CD44 promotes cell proliferation in non-small cell lung cancer. Oncol Lett 2018; 15: 5627-5633.
[Crossref] [Google Scholar] [PubMed]
- Ruiz EJ, Lan L, Diefenbacher ME, Riising EM, Da Costa C, Chakraborty A. JunD, not c-Jun, is the AP-1 transcription factor required for Ras-induced lung cancer. JCI Insight 2021; 6: e124985.
[Crossref] [Google Scholar] [PubMed]
- Hsu TI, Wang MC, Chen SY, Yeh YM, Su WC, Chang WC, Hung JJ. Sp1 expression regulates lung tumor progression. Oncogene 2012; 31: 3973-3988.
[Crossref] [Google Scholar] [PubMed]
- Cao LL, Song X, Pei L, Liu L, Wang H, Jia M. Histone deacetylase HDAC1 expression correlates with the progression and prognosis of lung cancer: A meta-analysis. Medicine (Baltimore) 2017; 96: e7663.
[Crossref] [Google Scholar] [PubMed]