Research Article - Biomedical Research (2018) Volume 29, Issue 4
Genetic diversity and antagonistic activity of plant growth promoting bacteria, isolated from tea-rhizosphere: a culture dependent study
Udaya Kumar Vandana1, Ankita Chopra1, Amarendranath Choudhury2, Dattatreya Adapa3 and Pranab Behari Mazumder1*
1Department of Biotechnology, Assam University, Silchar-788011, Assam, India
2Alumnus, Department Life Science and Bioinformatics, Assam University, Silchar-788011, Assam, India
3Department of Microbiology, Food Science and Technology, GITAM Institute of Sciences, GITAM University, Visakhapatnam, Andhra Pradesh, India
Accepted date: February 19, 2018
DOI: 10.4066/biomedicalresearch.29-18-428
Visit for more related articles at Biomedical ResearchAbstract
Microbial activity in root environment is responsible for plant nutrition, growth and defence. The objective of this study is to analyse the plant growth promoting and antagonistic activity rendered by the rhizospheric bacteria of tea plant. In this study, 292 bacterial isolates were screened for indole acetic acid (IAA) production, phosphate solubilisation, ammonia production, chitinase production and protease production. Among all the bacterial isolates, 58 isolates were able to elicit minimum four plant growth promoting rhizobacteria (PGPR) traits, which were further analysed quantitatively for hydrogen cyanide (HCN), siderophore production and antagonistic activity. Based on the plant growth promoting potential scores and principle component analysis, 12 samples were further screened for the study of salt tolerance and antifungal activity profile against tea fungal pathogens (Rhizoctonia solani, Fomes lamenensis, Corticium invisum). The identity the isolates was revealed by 16s rDNA sequence analysis. Six isolates (n63, k32, n61, 31k, n57, n56) showing efficient PGPR traits were evaluated for growth promotion studies on rice seedlings. Isolate n61 (Bacillus cereus) induced significant increase in root length (9.85 mm), shoot length (7.86 mm), germination percentage (92.44) and vigour index (1566.42) in rice seedlings. Fresh bio mass was significantly higher in 31k (Pseudomonas putida) (81.33 mg) followed by n61 (Bacillus cereus) (80.84 mg) and dry mass was higher in n63 (Bacillus pseudomycoides) (15.84 mg) followed by n61 (Bacillus cereus ) (15.82 mg) while compared with control and the other isolates selected for in vitro growth experiments. ANOVA analysis showed significant (P<0.05) increase compared with control, indicating that, bacterial isolates are potent for plant growth promotion and productivity.
Keywords
Plant growth promoting bacteria, 16S rDNA, Tea rhizosphere, Principle component analysis, Biocontrol
Introduction
Use of chemical fertilisers in excess is harmful to the soil and plant health. Extensive usage of chemical fertilisers causes damage to soil nutrients stability and biodiversity of rhizosphere in the long run. Recent reports showed that increased use of chemical fertilisers cause groundwater contamination, which in turn causes gastric cancer, goitre, birth malformations, hypertension, testicular cancer and stomach cancer [1,2]. Excessive air and water-borne nitrogen from fertilisers may cause hazardous effects to human health such as respiratory ailments [3,4], cardiac diseases, and several cancers [5,6]. Hence, alternative sustainable agricultural practices and biotechnological approaches to increase crop productivity, improve soil health and conserve biodiversity are in great demand [7].
Plant nutrition, growth and defence are associated with microbial activity present in root and surrounding soil-rhizosphere and such bacteria are specifically known as plant growth promoting rhizobacteria (PGPR) [8]. Plant growth promoting rhizobacteria (PGPR) influence the growth of plants either directly (production of plant growth regulators, improvement in plant nutrition, promote induce systemic resistance (ISR) or indirectly (production of metabolites such as antibiotics, hydrogen cyanide (HCN), siderophore) in rhizosphere [9]. In recent years, many studies on rhizosphere bacteria in different crops have been reported [10,11]. Studies on bacteria for plant growth promoting traits like, phosphate solubilisation, siderophore production, presence of nif H gene, chitinolytic activity and indole acetic acid (IAA) production, cellulase, protease, cyanic acid production and antibiotic production was reported on various crops and different soil ecosystems. This has increased the knowledge and importance of plant growth promoting bacteria and their role in sustainable agricultural development [11].
Tea (Camellia sinensis (L.) O. Kuntze) is one of the most important plantation crops in Assam, India. More than 50% of Tea is produced from this region of India [10]. Tea is the second most cheapest drinking beverage after water. Since the immortal time of human history, use of Tea has been mentioned in several ancient texts. Mainly, China is regarded as the origin of tea drinking and is also considered as a health-promoting habit [1,3-5]. Modern scientific understanding has supported the beneficial contribution of Tea through adequate experimental evidences. Cultivation of Tea plant is one of the oldest agricultural successes that are continuing till date. Studies have highlighted several protective contributions of Tea against deathly disease pathologies [5]. Recent studies have shown that, Tea drinking habit is having cancer-preventing potency and also assists to cope up with several cardiovascular and neurological complications. Undoubtedly, the role of polyphenols are determining in this regards. Though, several other phytocomponents are also having the potential to fight against oxidative stress and chronic inflammation [12] but the case of Tea is different as it is a well appreciated habit of daily life. Mainly, catechins and the aflavins are two major phenolic compounds which are known to provide all the protective effects of Tea.
Over the years, for increasing productivity there has been a continuous use of huge quantities of chemical pesticides and fungicides in tea plantations. Indiscriminate use of chemical fertilisers for long period of time reduced the soil health and tea productivity [13]. Hence, there is a need in tea industry for reducing the use of chemicals by supplementing with natural agents to increase productivity and control of diseases. Beneficial microbes like, Pseudomonas sp. [14], Bacillus spp. [15], Klebsiella spp. [16], Rhizobium [17], Azospirillum [18], Burkholderia sp. [19] have been reported in different crops like rice [20], green gram [21], wheat [22], chickpea [23], maize [24], black gram [25], barley [15], brassica [26], soybeans [27], sunflower [28], tomato [29] but very less research has been done on Tea PGPR. A large number of bacterial existences are found including nitrogen fixing bacteria in Tea soils of China [30]. Presence of PGRP in established Tea bushes in India was found by Pandey and Palni [31]. Indigenous plant growth promoting bacteria tend to give promising results in growth and production of different crops [32].This study aims to investigate the presence of dominant plant growth promoting bacteria and their biocontrol activity against common fungal pathogens in Tea rhizosphere of Assam, India.
Materials and Methods
Study site
Field surveys were carried out to collect the rhizospheric soil samples of different Tea varieties from different Tea gardens of Assam (India) namely Rose-Kandy Tea Estate, Heraband Tea Estate, Bongaigaon Tea Estate and Tea Research Station Karimganj (Tocklei).
Sample collection and isolation of rhizobacteria
The rhizospheric soil samples were collected from both nursery (9 months old to 1.8 years old) and adult (10-20 years) Tea plants. Root adhering soil was collected from nursery plants, and soil from a depth of 5 cm to 25 cm was collected from adult plants and placed in sterile polyethylene bags; brought to the laboratory within 24 h of collection and stored at 4°C for further isolation. Soil analysis was done using HiMedia (K-054) soil testing kit.
To isolate rhizobacteria, 1 gm of the rhizosphere soil sample was added to 9 ml of phosphate buffer (20 mM, pH 7.0) and incubated on a rotary shaker for 30 min at 150 rpm at 30°C. The processed soil samples were serially diluted up to 10-7. Sample was properly mixed by gentle shaking manually and spread plated on Nutrient agar medium, Tryptic soy broth, Kings B Medium and Burks N-free medium on three replicates and then incubated at 37°C for 3-5 days. A total of 292 colonies were isolated based on morphology and purified with repeated culturing and maintained in 30% glycerol at -80°C in ultra-low temperature freezer (Eppendrof, Germany) until next use [33].
Plant growth promoting traits
Phosphate solubilising activity: Rhizobacteria were determined quantitatively for phosphate solubilisation according to the method described by Nautiyal [34]. Bacterial strains were tested by plate assay using Pikovskya medium (HiMedia, India). Bacterial culture (5 µl) was placed on the plates and then incubated at 30°C for 7 days.
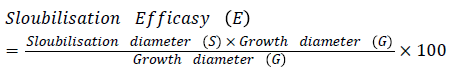
Indole acetic acid (IAA) production: Bacteria were grown at 37°C for 72 h in Lysogeny broth (LB) (100 μg ml-1, L-tryptophan supplemented) and centrifuged at 3000 g for 30 min. One ml of the supernatant was mixed with 4 ml of the Salkowski reagent (50 ml of 35% of Perchloric acid plus 1 ml of 0.5 M FeCl3). The intensity of the colour was measured at 530 nm and IAA standard curve was plotted using pure IAA (Sigma Aldrich, USA) [35].
Siderophore activity: Chrome azurol S (CAS) medium was prepared according to the method proposed by Pandey et al. [32]. Siderophore detection was done by following O-CAS, a fast and universal method for siderophore detection described by Ads et al. [37], Cankiliç et al. [38], and Pérez-Miranda et al. [39]. Quantitative estimation of siderophore was done by using CAS shuttle assay. Briefly, 0.5 ml of culture supernatant was added into 0.5 ml of CAS solution. Change in the colour indicated siderophore production. OD was measured at 630 nm after 20 min of incubation. Uninoculated broth supernatant mixed with CAS solution served as control.
Siderophore quantity percentage was calculated using the following formula-
Siderophore quantity percentage=Ac - As/Ac × 100
Where, Ac: Absorbance of Control; As: Absorbance of Sample
HCN production: Hydrogen Cyanide (HCN) production was assessed using nutrient agar supplemented with 0.44% (w/v) of glycine. The agar surface, streak-inoculated with overnight culture, was over laid with a Whatman filter paper (no. 1) soaked in 2% (w/v) sodium carbonate in 0.5% (v/v) picric acid and incubated at 30°C for 72 h. Change in colour of the filter paper from yellow to orange, red or brown indicated lesser, moderate or higher levels of HCN production, respectively [40]. Quantitative analysis was done by hanging the filter paper strips amended with picric acid and sodium bicarbonate. After incubating the culture growing tube at 30°C for 7 days, strips kept in double distilled water (10 ml) and change in the colour was observed at 625 nm [41].
Chitinase activity: Chitinase activity was determined by the method described by Chernin et al. [42]. Colloidal chitin (0.2%) was added in trypticase soy agar (TSA). N-Acetyl-D-Glucosamine was measured calorimetrically at 585 nm by using colorimetric agent di-methyl amino benzaldehehyde [43].
Protease activity: The isolates were screened for the production of protease by streaking them on Skim milk agar [44].
Ammonia production: Bacterial isolates were tested for the production of ammonia in peptone water. Development of brown to yellow colour was a positive test for ammonia production [45].
Salt tolerance: Tolerance of the isolates against salinity was examined by observing the growth on nutrient agar medium composed with final concentrations of NaCl (3, 5, 8, 10% (w/v)). The plates were incubated for 48-72 h at 28 ± 2°C. Same experiment was carried out with NaCl amended broth. Cultures with different concentrations (3, 5, 8, 10% (w/v) of NaCl amended broth was incubated at 35°C for 7 days and OD was measured at 600 nm [46].
Antagonistic activity: Selected rhizobacteria antagonistic activity against Rhizoctonia solani, Corticium invisium, Fomes lamonensis was determined by employing a dual culture technique [35]. The radii of the fungal colony towards and away from the bacterial colony were measured [47].
Antifungal activity was depicted by using the formula: Dc - Dt/Dc × 100
Where, Dc: Diameter of fungal growth in control; Dt: Diameter of maximum fungal growth in test sample
Seed germination: Seeds were surface-sterilized using 2% (v/v) sodium hypochlorite for 2 min, followed by repeated wash with sterile double distilled water and air-dried under laminar air flow. The sterile seeds were soaked in bacterial suspension (108 CFUs/ml) for 6 h on shaking incubator at 30°C., sterile double distilled served as a control [48].
In vitro plant growth promotion assay: Germination test was carried out by the paper towel method according to the standard procedures of International Seed Testing Association (ISTA). Seedling vigour was analyzed as per the method described by Appa Rao et al. [49]. The vigour index (VI) was calculated using the formula:
V=(mean root length + mean shoot length) × (% germination).
16SrDNA Sequencing and phylogenetic analysis: Extraction and purification of the genomic DNA was done using Quiagen DNA isolation kit. 16S rDNA gene fragment was amplified using a master cycler gradient thermal cycler (Applied Biosystem, USA) using universal primers (27F, 1492R). The PCR fragments were sequenced using ABI PRISM 3730 XL DNA sequence, Assam University, Department of Biotechnology (Assam, India). The DNA sequences obtained were compared with 16S rDNA gene sequences in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) search algorithm according to the method proposed by Altschul et al. and Arora et al. [50,51] to determine similarity of the partial 16S rDNA sequences. The nucleotide sequences were submitted to the NCBI database and the accession numbers were obtained.
Statistical analysis
Statistical analysis on the effects of the bacterial strain on PGP traits (IAA and siderophores production, phosphate solubilisation, chitinase, protease, HCN, ammonia production and antifungal activity) was performed in Excel 2010. The data of in vitro plant growth experiments was performed for analysis of variance (ANOVA). Principal component analysis (PCA) was performed by using Past version 2.07 software to determine correlations and variation between different PGP activities.
Results
The descriptive analysis of the study outcome has been presented (Figures 1-5 and Tables 1-4). A total of 292 bacteria were isolated and purified from various soil samples from different locations. Soil pH, texture, contents namely phosphate, potassium, ammonical nitrogen content, nitrate nitrogen, and oxidizable organic carbon has been summarized in Table 1.
| Type of Soil Sample | Kalain | Karimganj | Heraband | Rosekandy |
|---|---|---|---|---|
| pH | 5.38 | 4.58 | 5.13 | 4.92 |
| Phosphate Content | Nil | Nil | Nil | Nil |
| Potassium Content | Very High | Very High | High | Very High |
| Ammoniacal Nitrogen | Low (About 15) | Low | Low | Low |
| Nitrate Nitrogen | Very Low (About 04) | Very Low | Very Low | Very Low |
| Oxidizable Organic Carbon | 0.300-0.500 (Medium Low) | 0.300-0.500 (Medium Low) | 0.505-0.750 (Medium) | 0.76 |
Table 1: Physico-chemical properties of different soil samples.
| SKIM | CHITIN | PVK | IAA | AM | ANT | SID | HCN | |
|---|---|---|---|---|---|---|---|---|
| n6 | ++ | ND | +++ | ++++ | ++ | ND | + | ND |
| n7 | ++ | ND | +++ | +++ | + | ND | ND | ND |
| n8 | ++ | ND | +++ | ++ | + | ND | ND | + |
| n11 | + | ++ | + | ++ | + | + | + | ND |
| n12 | ++ | + | + | ++ | ++ | + | ND | + |
| n14 | + | + | ND | ++ | ++ | ND | + | ND |
| n15 | ND | ND | ++ | ++ | ++ | ND | ND | + |
| n2ND | ++ | + | + | ++ | ++ | ND | ND | ND |
| n21 | + | ND | +++ | ++ | ++ | ND | ND | ND |
| n23 | +++ | ND | + | ++ | ++ | ++ | ND | ND |
| n24 | +++ | ND | + | ++ | ++ | ND | ++ | + |
| n3ND | +++ | + | + | + | ++ | ND | ND | ND |
| n13 | + | + | ++ | + | +++ | ND | ND | ND |
| nt6 | + | + | + | + | ++ | ND | ND | ND |
| nt7 | + | ND | + | + | ND | ND | +++ | ND |
| nt9 | ++ | ND | ++ | + | +++ | + | ++ | ND |
| nt17 | ++ | ++ | + | + | ND | ND | ND | + |
| nt2ND | ++ | + | + | ++ | ND | ND | ND | + |
| nt24 | + | ++ | + | ND | ND | ++ | ++ | ND |
| nt62 | ++ | + | +++ | ++++ | + | ND | ND | ND |
| m61 | + | ++ | ++ | + | + | ND | ND | ++ |
| m58 | + | +++ | + | + | + | + | ND | + |
| mt7 | ++++ | ND | + | + | + | ND | ND | ND |
| mt2 | ++++ | ND | + | ++ | + | ND | + | ND |
| mt49 | ND | + | ++ | +++ | + | ND | + | ND |
| mt1ND | + | ND | ++ | +++ | + | ND | ND | ND |
| mt21 | ++ | ++ | ++ | + | + | ND | ND | ND |
| mt23 | ND | +++ | ++ | + | + | ND | ND | + |
| mk4 | + | ND | + | ++ | +++ | ND | ND | ND |
| mk5 | +++ | ND | + | + | + | ND | + | ND |
| mk6 | + | ND | + | + | +++ | ND | + | ND |
| mk8 | + | ND | +++ | + | + | ND | ++ | ND |
| mk15 | + | ND | + | + | ++++ | ND | ++ | ND |
| mk24 | + | + | + | + | ND | ND | ++ | +++ |
| mk28 | ND | + | ++ | ++ | +++ | ND | ND | +++ |
| mk37 | + | + | ND | + | +++ | + | + | + |
| mk43 | + | ND | + | ++ | ++ | + | ND | + |
| ml32 | + | ++ | + | ND | + | + | ND | ND |
| ml2ND | + | + | ++ | + | + | ND | ND | ND |
| ml11 | + | ND | ++ | + | + | ND | ND | ND |
| mb2 | ND | + | ++++ | ++ | + | + | + | + |
| mb8 | ND | ++ | ++++ | + | + | ND | ND | ND |
| mb9 | + | ND | ++++ | + | + | + | ND | ND |
| mb16 | ND | ND | + | + | + | ++ | ND | ND |
| mb18 | + | ++ | ND | + | + | ++ | ND | ND |
| mb19 | ND | ++ | +++ | + | + | ND | +++ | ++ |
| N51 | ++ | + | ++ | ++++ | ++ | ++ | ++ | ND |
| N61 | +++ | ++ | +++ | ++++ | +++ | +++ | + | + |
| 31k | +++ | ND | +++ | +++ | + | ++ | +++ | +++ |
| N35 | + | ++ | +++ | +++ | ++ | +++ | ND | ND |
| N63 | ++++ | + | ++ | +++ | +++ | ++ | + | ND |
| K35 | +++ | +++ | ++ | ++ | ++ | ++ | +++ | + |
| N4 | ++ | + | +++ | ++ | + | +++ | + | + |
| 3N | +++ | ++ | ++ | + | + | +++ | + | + |
| N17 | ND | ++ | +++ | ++++ | + | ++ | ++ | ND |
| N56 | + | +++ | +++ | ++++ | +++ | + | ++ | ++ |
| N57 | ++ | + | ++++ | +++ | ++ | +++ | + | ++ |
| K32 | ++ | + | +++ | ++++ | +++ | ++ | +++ | ++ |
| +=Less Activity; ++=Good; +++=Very Good; ++++=Excellent activity Protease: 2-7 mm=+; 7-15 mm=++; 15-20 mm=+++; 20-25 mm=++++; IAA (ug/ml): 5-15=+; 15 -25=++; 25-35=+++; 35 and above=++++; Phosphate (SU): 100-120=+; 120-140=++; 140-160+++; 160 and above ++++ SKIM: Protease Production; CHITIN: Chitinase production; PVK: Phosphate solubilisation; IAA: IAA production; AM: Ammonia Production; ANT: antagonistic activity; SID: Siderophore production; HCN: HCN production |
||||||||
Table 2: Plant growth promoting attributes of selected isolates.
| Isolate | Shoot length (mm) | Root length (mm) | Fresh weight (mg plant-1) |
Dry weight (mg plant-1) |
Vigour index | Germination (%) |
|---|---|---|---|---|---|---|
| Control | 4.22 ± 0.76 | 4.74 ± 0.79 | 60.34 ± 12.16 | 14.73 ± 3.22 | 805.00 | 89.77 |
| n63 | 7.35 ± 1.55 | 5.97 ± 1.23 | 63.82 ± 5.23 | 15.84 ± 1.58 | 1202.96 | 90.22 |
| k32 | 7.08 ± 0.71 | 6.97 ± 2.17 | 64.48 ± 9.80 | 14.93 ± 2.72 | 1225.36 | 87.11 |
| n61 | 7.85 ± 1.05 | 9.08 ± 1.41 | 80.84 ± 13.26 | 15.82 ± 1.38 | 1566.42 | 92.44 |
| 31k | 6.98 ± 1.28 | 7.35 ± 1.02 | 81.33 ± 9.37 | 14.95 ± 1.88 | 1179.43 | 82.22 |
| n57 | 6.45 ± 0.92 | 6.31 ± 0.94 | 63.46 ± 14.15 | 14.82 ± 1.87 | 1163.18 | 92.11 |
| n56 | 7.5 ± 1.56 | 6.31 ± 0.86 | 70.45 ± 8.67 | 15.10 ± 1.73 | 1252.2 | 90.66 |
Table 3: Effect of selected isolates on rice seedling growth promotion.
| Sample code | Antagonism | Gene bank accession no. |
Nearest homolog (>99% similarity) |
Salt Tolerance | |||||
|---|---|---|---|---|---|---|---|---|---|
| C. invisium | F. lamonensis | R. solani | 3% | 5% | 8% | 10% | |||
| N51 | ++ | + | ++ | KM 403654 | Bacillus cereus | + | + | + | + |
| N61 | + | +++ | KM 502964 | Bacillus cereus | + | + | + | + | |
| 31k | ++ | ++ | KR 049181 | Pseudomonas putida | + | + | + | ||
| N35 | +++ | KR 066418 | Bacillus cereus | + | + | ||||
| N63 | + | ++ | KR 517569 | Bacillus pseudomycoides | + | + | + | +* | |
| K35 | ++ | KM 820830 | Pseudomonas stutzeri | + | + | + | + | ||
| N4 | + | +++ | +++ | KM 585688 | Pseudomonas stutzeri | + | + | + | |
| 3N | +++ | +++ | KR 013205 | Pseudomonas stutzeri | + | + | + | + | |
| N17 | ++ | KM 388541 | Bacillus methylotrophicus | + | + | + | + | ||
| N56 | + | + | KM 658173 | Bacillus cereus | + | + | |||
| N57 | ++ | +++ | KM 375933 | Bacillus pseudomycoides | + | + | + | + | |
| K32 | + | ++ | KM 624614 | Pseudomonas stutzeri | + | + | + | + | |
| HCN: 0.02, ++ 0.02-0.04, +++ 0.04-0.06, ++++ 0.06 (OD) Chitins: + 70, ++ 70-85, +++ 85-95, ++++ 95 (U mL-1) Antagonistic activity: 40%-50%=+, 50-64=++, 65-75=+++ Siderophore: 10-20=+, 20-35=++, 35-50=+++, 50-65=++++ Ammonia faint yellow=+; Deep Yellow=++; Feeble brown=+++; Deep reddish brown=++++ |
|||||||||
Table 4: Molecular identification of PGPR isolates with their NCBI accession numbers and their antagonistic activity profiles.
Plant growth promoting traits
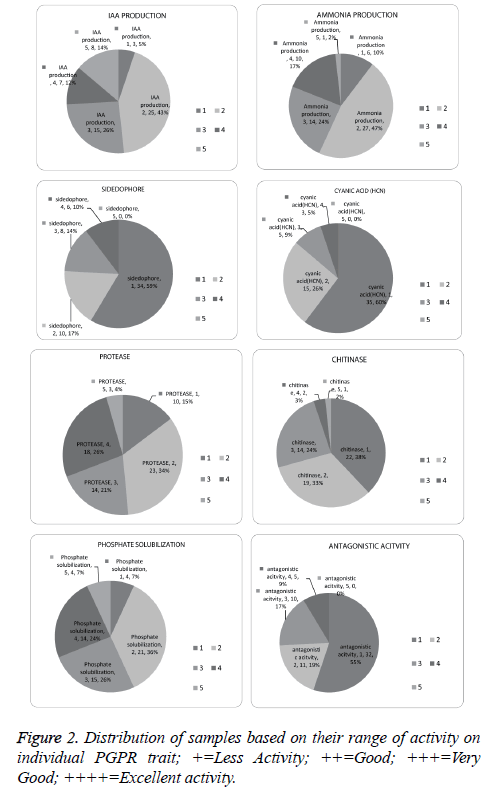
Screening for protease, chitin, phosphate solubilisation, IAA and ammonia was conducted for 292 bacteria, among these 67.4% were found positive for protease, 23.6% for chitin, 54.1% for phosphate solubilisation, 68.4% for IAA and 60.9% for ammonia production (Figure 1).
Fifty eight selected isolates possessing more than 4 PGP traits were considered for further level of screening steps for plant growth promotion attributes which include antagonistic activity, HCN production, siderophore production, etc. (Table 2).
Phosphate solubilisation efficiency (PSE) was calculated after seven days of incubation. Fifty-five isolates were found positive for phosphate solubilisation. Among all the isolates, mb2 was found to show more efficiency. Four isolates (mb2, mb8, mb9, and n57) produced more than 160 units of solubilisation.
IAA production is the most predominant trait among the isolates in our study. Out of 58 samples, 56 samples produced IAA. Isolate N61 produced highest IAA (74.53 µg ml-1), 20 isolates produced more than 10 µg ml-1, ammonia production was found in 53 isolates and n63 showed highest (reddish brown) Ammonia production (Figure 2).
A total of 58 isolates were investigated for siderophore production and 24 isolates were confirmed by orange halo. Further quantitative analysis was performed for samples which were positive. It was found that 31k, nt7, nt24, mb19, k35 were producing sufficient quantity of siderophore with more than 40% siderophore production units (SU) (Figure 2).
Some bacteria promote plant growth through antagonistic activity against phytopathogens through the production of hydrolytic enzymes. Major fungal cell wall components are made up of chitin and beta-glucan, thus chitinase production is performed [52]. Thirty six isolates produced chitinase in which K35 produced the maximum amount of chitinase; n56, mt23, m58 were found to be good in terms of efficiency in chitinase production.
Protease production helps plant in growth promotion indirectly by major number of bacteria. Forty-four isolates were found positive for protease production. Among all the isolates mt7, mt2, N63 were found equally efficient (Figure 2).
HCN production was found to be the least expressed trait among the isolates in this study. Only Twenty-three isolates out of 58 isolates were positive. Isolates 31k, mk28, and mk24 showed higher levels of HCN production (Figure 2).
Salt tolerance of isolates was identified by the presence and absence of growth on the agar plate after 7 days of incubation. In broth OD, above 0.5 was considered as positive. All the 12 isolates were found positive for 3%, 5% salt where ten isolates were positive for 8% and 8 isolates out of 12 were positive for 10% salt stress.
A total of 58 isolates were screened for in-vitro antagonistic activity, out of which 26 isolates were found positive against Fomes lamenensis. Twelve selected isolates were found best in plant growth promoting attributes with antagonistic activity and further screened with another two fungi Corticiu mininvisium and Rhizoctonia solani.
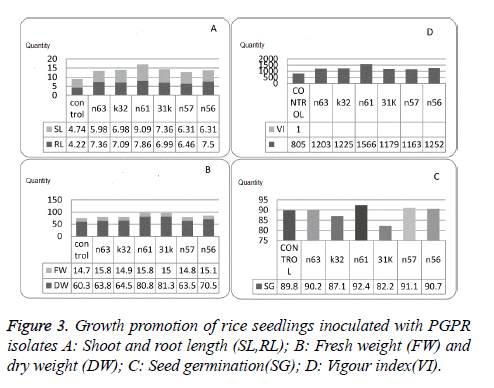
In vitro plant growth promotion assay
All the selected isolates showed significant increase in growth and germination percentage. Isolate n61 was found to be effective growth promoter as it showed 92.4% germination and 1566.4 vigour index which was higher than untreated seeds (Figure 3, Table 3).
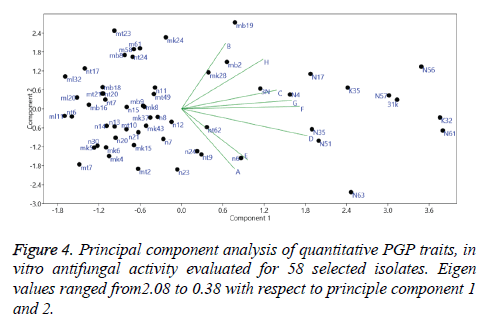
Principle component analysis
Principal component analysis (PCA) was performed based on the amounts of PGP activities expressed by the strains. Result of which have shown 27.6% to 4.8% variance and Eigen values ranging from 2.08 to 0.38 among the first and second PCA, respectively. Analysis of the data demonstrated correlation between PGP traits concerning behaviour of strains and their expression (Figure 4).
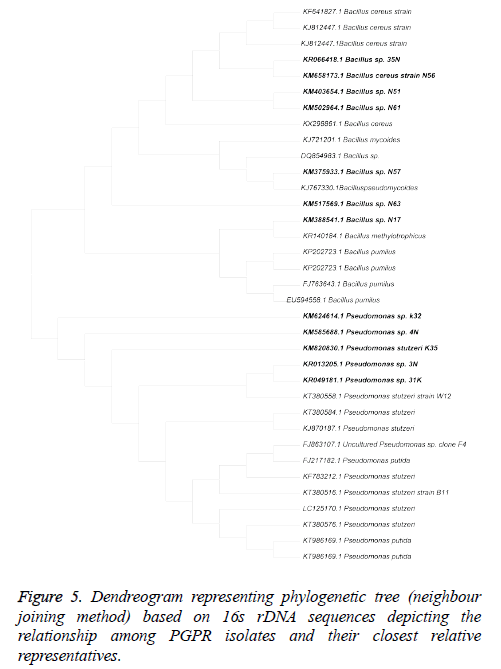
16SrDNA Sequencing and Phylogenetic analysis
The results of 16S rDNA sequencing analysis showed that out of 12 efficient isolates belongs to 2 different genera i.e. Pseudomonas (4) and Bacillus (8). The evolutionary history was inferred using the Neighbour-Joining method (Figure 5). The optimal tree with the sum of branch length=0.34624451 is shown. The evolutionary distances were computed using the Kimura 2-parameter method. All positions containing gaps and missing data were eliminated. There were a total of 1093 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 (Table 4).
Discussion
Rhizosphere is a resourceful and robust ecosystem which facilitates continuous microbe-plant interaction for reining plant growth. In the present investigation, 292 isolates were screened for plant growth promoting capability out of which 58 isolates showing promising plant growth promotion and antifungal activity, were selected for further experiments. Twelve isolates were found to be efficient in plant growth promoting activity and antifungal activity, further characterisation by 16S rDNA sequencing revealed that isolates showed more than 99% similarity with Bacillus methylotrophicus (1), Bacillus mycoides (1)., Bacillus pseudomycoides (2), Bacillus cereus (4), Pseudomonas stutzeri (4). Six best isolates based on their PGP efficiency and growth promotion strains were used for in-vitro growth promoting experiments i.e. seed bacteriazation and isolates effect on seed germination and seed vigour index. All the bacteria depicted their significance on the growth promotion of rice seedlings and germination. In this study Bacillus and Pseudomonas isolates were found predominant and effective plant growth promoters. Selected isolates showed PGP activities with variability in the percentage of positive isolates for individual traits.
Phosphate solubilising bacteria play an important role in plant nutrition by making the insoluble phosphorous available for plant intake. 94% of isolates were able to solubilise phosphate (Ca3PO4) indicating that soil is rich in phosphate solubilising bacteria. Various studies on phosphate solubilization illustrated decrease in PH and production of extracellular organic indicate phosphate solubilisation [53,54]. In our study it was found that soil is slightly acidic and available phosphate is moderate. Genes (gcd and pqq) involved in the production of glucose dehydrogenase and PQQ fundamentally affected the phosphate solubilization ability of Pseudomonas fluorescensF113. This demonstration by Miller et al. [55] provided a greater understanding of the mechanisms in solubilisation of inorganic phosphate.
Auxins play a pivotal role in root growth and development [56]. Among all the isolates, n61, 31k, k32 showed higher IAA production and increased root length. It may in return benefit plant in higher nutrient intake [57]. Similar results were found in different studies on PGPR. It was also observed that n56 isolate which was found to produce higher amount of IAA could not influence the root length like the other samples with higher IAA production. Although IAA is mainly known to induce lateral root formation and development, recent study found negative correlation between root length and IAA production and positive correlation with root biomass and growth [57]. In our study, we found that IAA production was the most (56/58 samples) expressed among all PGP traits tested.
Ammonia producing bacteria converts organic nitrogen to ammonical nitrogen which leads to increase in soil nitrogen content. We were able to find that after IAA production and phosphate solubilisation, ammonia production was found predominant among the selected isolates. Alkalisation caused by ammonia production suppresses the growth of some fungal pathogens which indirectly promotes plant growth [58]. Majority of the isolates produced fair range of ammonia i.e. more than deep yellow, only one isolate n63 produced reddish brown which indicates higher range of ammonia production. But in our study, we observed that isolate n63 showed lesser seed germination among the selected isolates in in-vitro plant growth promotion. Similar results were demonstrated on S. odorifera 4Rx13 in compartmented petri dishes caused subsequently reduced growth of A. thaliana due to alkalization of the neighbouring plant medium. Concentration of ≤ 1 µmol had less impact on growth but ≥ 2.5 µmol showed higher rate of growth inhibition. Weise et al. study supported that higher amount of ammonia production will effect plant growth [59].
Some bacteria promote plant growth by producing low density iron chelator called siderophore. These molecules can help plant growth in two different ways. One is directly facilitating insoluble iron to plant and the other is antagonism against fungal pathogens by making iron unavailable for fungal minimum nutrition [60]. Reports found that siderophore producing bacteria will contribute by increasing dry weight and green foliage. It was also reported to create healthy soil that suppress fungal growth [61,62]. In our study, 43% of selected bacteria found positive for siderophore production. This indicates that a decent number of bacteria were contributing soil health of this region. Among the 12 best isolates, 31k, k35, k32 found to be the best siderophore producers. Interestingly all the 3 best siderophore producers belong to the Pseudomans spp. Although Bacillus spp. has good number of siderophore producers, in our study, Pseudomonas spp. were found to have more efficiency. Efficient siderophore producing isolates showed good results in plant dry weight over the control and other tested samples. Studies found that PH plays a conductive role in phosphate solubilisation and siderophore production. As Tea soils are acidic it might have contributed for siderophore production. Recent findings on siderophores demonstrated that iron availability can also regulate siderophore production which in turn depends upon soil pH [63,64].
Low molecular weight substance like HCN and lytic enzymes like chitinase, protease was found to contribute plant growth promotion [65]. Kumar et al. demonstrated that chitinase and β-1,3-glucanase enzymes enhances growth and biocontrol of Cajanus cajan L. Var. In the current study, we found 62% isolates exhibited chitinase production. k35 and n56 isolates found to have higher activity of chitinolysis. It was also observed that the isolates with higher amount of chitinase were efficient suppressors of all the three fungi with good inhibition percentages in dual culture assay. A study on Pseudomonas spp. demonstrated that it will use chitin as energy source [66]. Similar observation was demonstrated by Kishore et al. [67] on partially purified chitin from the isolate, able to control Botrytis grey mold disease. Considering the above results, our study could be added to the literature on chitinolytic bacteria biocontrol.
Protease is another key hydrolytic enzyme which can contribute to biocontrol activity against phytopathogens. Demonstrations on supplemented protease with heat treated culture supernatant depicted the proteolytic bacteria and its biocontrol activity on R. Solani [52]. Similar studies on Stenotrophomans maltphilia biocontrol activity against Ralstonia solanasearum illustrated protease genes expression (prt genes) correlated with biocontrol activity [68]. In our study, n63, mt7, mt2 showed higher proteolytic zones. Out of all selected isolates, 84% of isolates were found to have proteolytic activity. All the 12 selected isolates were possessing proteolytic activity except n17, but still it was able to suppress fungal growth of Corticium invisium and Fomes lamenensis. It may be due to chitinase or siderophore activity of that isolate.
Inorganic compounds such as HCN, H2S, Nitric oxide are recently referred as bacterial volatile compounds (BVC) rather than volatile organic compounds (VOC) [69]. HCN producing isolates have shown antagonistic activity against pathogenic fungi. It inhibits metalloenzyme cytochrome oxidase activity and thus plays a key role in biocontrol activity [70]. HCN producing Pseudomonas spp. have been well documented in the literature with reference to their biocontrol activity and their correlation with Phl (phenazine 1- carboxylic acid). In our study, HCN production was found in lesser extent when compared with other traits. Only 39% isolates exhibited HCN production. 31 K i.e Pseudomonas spp. showed highest HCN production. Out of 12 best isolates, 8 were found positive and in which 4 isolates found fair and 3 isolates found good and 1 isolate (31k) showed excellent activity. It was also found in our study that 31k did not show much effect on root growth, supporting the work demonstrated by Siddiqui [71]. Principle component analysis on plant growth promoting traits showed significant variance among the samples and correlation was observed among some of the traits.
Salinity stress is one of the major abiotic stresses which cause yield reduction in many crops [72-75]. Application of PGPR is been reported as viable alternative to reduce abiotic stress on plants by maintaining osmotic balance. In the present study, isolates (N61, N51, K35, N63, 3N, N17, N57and k32) grown under different salt concentrations indicated their resilience under unfavourable conditions. Pseudomonas and Bacillus spp. grown at high salt concentration were demonstrated as plant growth promoters under stress conditions in earlier reports [76,77]. However, our study found the presence of salt tolerance in Pseudomonas and Bacillus spp. along with growth promoting traits.
Capability of subduing pathogenic fungal growth renders plant growth promoting activity. Bacillus and Pseudomonas spp. were well ascertained as biocontrol agents in several works. In our current study, we found Bacillus sps showed relatively more biocontrol activity over Pseudomonas spp. n61, n35,4N, n57 were able to suppress the growth of all three fungi with considerable zone of inhibition. Interestingly, n56 which was found to be a good siderophore, HCN and chitin producer was able to suppress only one fungi Rhizoconita solani. It could not control the growth of Corticium invisium and Fomes lamanensis. Available reports show that chitinase, siderophore producers were able to control Rhizoctonia solani growth. We may draw the conclusion from the current study that best antagonistic isolates (n61,n35,4N,n57) which were able to suppress the growth of 3 fungi were dependent on protease production along with siderophore and chitin with minor exceptions. We also found that dominant Tea rhizosphere bacteria Pseudomonas and Bacillus as plant growth promoters and biocontrol agents. They may be added to available literature of Tea rhizosphere microbes.
In vitro growth promotion experiments indicated significant variability when compared control with selected isolates and also among the selected isolates. Seed treated with isolate n61 showed significant increase in root length (9.85), shoot length (7.86), germination percentage (92.44), and vigour index (1566.42) followed by n56 vigour index (1252.20) and germination percentage (90.66). Fresh bio mass is significantly higher in 31k (81.33) followed by n61 (80.84) and dry mass is higher in n63 (15.84) followed by n61 (15.82) over the control and the other isolates selected for in vitro growth experiments. PGPR with higher IAA production, extracellular metabolite production and volatile production tend to promote growth to maintain plant microbe interaction.
Previous reports found the dominance of Bacillus and Pseudomonas in Tea rhizosphere. In the current study, we found that they elicited the PGPR traits and antifungal activity to a considerable extent. Hence they are potential plant growth promoters.
Conclusion
Reports regarding the inconsistency of microbial fertilizers on fields have been a part of discussion since many years. Major reason can be the origin of isolation and place of inoculation. In many cases, isolates cannot survive or cannot establish their dominant plant growth promoting traits due to their specificity. Native strains used as bio inoculants found to give significant growth promotion because of their host rhizosphere specificity. Bacillus spp., because of their spore formation, susceptibility to various climatic conditions; and Pseudomonas spp. for their fastidious colonisation behaviour and antagonistic activity towards potential fungal pathogens will be a viable alternative in organic farming of Tea.
Acknowledgement
Authors are thankful to Department of Biotechnology (DBT), Delhi, India for the financial support.
References
- Kristensen P, Andersen A, Irgens LM, Bye S, Vagstad N. Testicular cancer and parental use of fertilizers in agriculture. Cancer Epidemiol Biomarkers Prev 1996; 5: 3-9.
- Rai N, Ashiya P, Singh Rathore D. Comparative study of the effect of chemical fertilizers and organic fertilizers on Eisenia foetida. Int J Innov Res Sci Eng Technol 2007; 3297: 12991-12998.
- Nian L, Liangyi X, Gang J. Analysis on constituents and drug resistance of pathogenic bacteria causing chronic obstructive pulmonary disease. Biomed Res 2017; 28: 8939-8942.
- Amoli RI, Nowroozi J, Sabokbar A. Isolation of Stenotrophomonas maltophilia from water and water tap. Biomed Res 2017; 28: 8660-8664.
- Wood SL, DeClerck F. Ecosystems and human well-being in the sustainable development goals. Front Ecol Environ 2015; 13: 123.
- Li X, Zhu Y. The value of monocyte volume index in predicting post-surgical bacterial infection. Biomed Res 2017; 28: 8996-8999.
- McDaniel MD, Tiemann LK, Grandy AS. Does agricultural crop diversity enhance soil microbial biomass and organic matter dynamics? A meta-analysis. Ecol Appl 2014; 24: 560-570.
- Hartmann A, Rothballer M, Schmid M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant and Soil 2008.
- Bloemberg GV, Lugtenberg BJJ. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 2001; 4:343-350.
- Saharan BS, Nehra V. Plant growth promoting rhizobacteria: A critical review. Life Sci Med Res 2011; 21: 1-30.
- Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 2009; 63: 541-556.
- Choudhury A, Chakraborty I, Banerjee TS, Vana DR, Adapa D. Efficacy of morin as a potential therapeutic phytocomponent: Insights into the mechanism of action. Int J Med Res Health Sci 2017; 6: 175-194.
- Chakraborty U, Chakraborty B, Chakraborty A, Sunar K, Dey P. Plant growth promoting rhizobacteria mediated improvement of health status of tea plants. Indian J Biotechnol 2013; 12: 20-31.
- Ahemad M, Khan MS. Effect of fungicides on plant growth promoting activities of phosphate solubilizing Pseudomonas putida isolated from mustard (Brassica compestris) rhizosphere. Chemosphere 2012; 86: 945-950.
- Canbolat MY, Bilen S, Cakmakci R, Sahin F, Aydin A. Effect of plant growth-promoting bacteria and soil compaction on barley seedling growth, nutrient uptake, soil properties and rhizosphere microflora. Biol Fertil Soils 2006; 42: 350-357.
- Ahemad M, Khan MS. Effects of insecticides on plant-growth-promoting activities of phosphate solubilizing rhizobacterium Klebsiella sp. strain PS19. Pestic Biochem Physiol 2011; 100: 51-56.
- Ahemad M, Khan MS. Toxicity assessment of herbicides quizalafop-p-ethyl and clodinafop towards rhizobium pea symbiosis. Bull Environ Contam Toxicol 2009; 82: 761-766.
- Rodrigues E, Rodrigues L, de Oliveira A, Baldani V, Teixeira K, Urquiaga S, Reis V. Azospirillum amazonense inoculation: effects on growth, yield and N-2 fixation of rice (Oryza sativa L.). Plant Soil 2008; 302: 1-2.
- Guo JK, Ding YZ, Feng RW, Wang RG, Xu YM, Chen C, Wei XL, Chen WM. Burkholderia metalliresistens sp. nov., a multiple metal-resistant and phosphate-solubilising species isolated from heavy metal-polluted soil in Southeast China. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol 2015; 107: 1591-1598.
- Mirza MS, Mehnaz S, Normand P, Prigent-Combaret C, Moënne-Loccoz Y, Bally R, Malik KA. Molecular characterization and PCR detection of a nitrogen-fixing Pseudomonas strain promoting rice growth. Biol Fertil Soils 2006; 43: 163-170.
- Wani PA, Khan MA, Zaidi A. Effect of metal tolerant plant growth promoting Bradyrhizobium sp. (vigna) on growth, symbiosis, seed yield and metal uptake by greengram plants. Chemosphere 2007; 70: 36-45.
- Khalid A, Arshad M, Zahir ZA. Screening plant growth-promoting rhizobacteria for improving growth and yield of wheat. J Appl Microbiol 2004; 96: 473-480.
- Verma JP, Yadav J, Tiwari KN, Jaiswal DK. Evaluation of plant growth promoting activities of microbial strains and their effect on growth and yield of chickpea (Cicer arietinum L.) in India. Soil Biol Biochem 2014; 70: 33-37.
- -->