Research Article - Biomedical Research (2017) Volume 28, Issue 14
Comparison of α-synuclein gene methylation in peripheral blood mononuclear cells of Uighur and Han patients with sporadic Parkinson's disease
Qin Luo1#, Huan Xia2#, Yalou Zhang3, Guolei Cao1 and Xinling Yang4*1Department of General Medicine, the Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, PR China
2Department of Nuclear Medicine, the Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, PR China
3Department of Histology and Embryology, Basic Medical College of Xinjiang Medical University, Urumqi, PR China
4Department of Neurology, the Second Affiliated Hospital of Xinjiang Medical University, Urumqi, PR China
#These authors contributed equally to this study
- *Corresponding Author:
- Xinling Yang
Department of Neurology
The Second Affiliated Hospital of Xinjiang Medical University, PR China
Accepted date: June 26, 2017
Abstract
This study aims to investigate the correlations between the methylation level of the Parkinson's Disease (PD)-susceptibility α-synuclein gene (SNCA) intron region 1 and the PD incidences in Uighur and Han ethnic groups. The Peripheral Blood Mononuclear Cells (PBMCs) of 40 patients (19 Uighur and 21 Han) with Early-Onset Parkinson's Disease (EOPD) and 40 normal people (19 Uighur and 21 Han, ≤ 50 y old) were isolated by density gradient centrifugation, and the methylation level of SNCA gene intron region 1 was then detected using the bisulfite Polymerase Chain Reaction (PCR) sequencing method. The comparisons between the EOPD group and the ≤ 50 y old control group, between the Han-EOPD group and the Han-control group, between the Uygur-EOPD group and the Uygur-control group, between the male EOPD group and the male control group, between the female PD group and the female control group, and between the male EOPD group and the female EOPD group all showed no significant difference in the average methylation level of the SNCA gene intron region 1 (P>0.05). The methylation level of the SNCA gene intron region 1 might not be associated with the EOPD incidence in Uighur and Han patients.
Keywords
Parkinson's disease, Susceptibility a-synuclein gene, Methylation.
Introduction
Parkinson's disease (PD) is a degenerative disease of the nervous system, mainly occurring in the elderly with long pathogenic path and high morbidity, and it’s the second most common neurodegenerative disease after Alzheimer's disease [1]. The etiology of PD is multifactorial and might involve in protein misfolding and aggregation [2], mitochondrial dysfunction [3], oxidative stress [4], or epigenetics [5]. DNA methylation is one of the most important epigenetic mechanisms, and studies in recent years suggest that PD might be related with the methylation. Jowaed et al. and Matsumoto et al. [6,7] found almost at the same time that the methylation levels of SNCA gene intron 1 in the cerebral substantia nigra, putamen, and cortical region of PD patients were decreased significantly, and the upregulation of certain genes were also accompanied during this process, resulting in the formation of insoluble Lewy bodies from a-synuclein via accelerating its aggregation and slowing down its degradation. However, Richter et al. [8] reported no significant difference in the methylation status of the peripheral blood leukocytes between PD patients and normal people, so no clear link between the methylation level of SNCA intron 1 and PD onset could be drawn, indicating there still exists controversies and needs further investigations about the correlations between the DNA methylation and PD. In Xinjiang, Uighur and Han people have different genetic backgrounds, and it has not been reported about whether there exists certain difference in the SNCA gene methylation between these two nationalities. This study started from another angle, namely the methylation of SNCA gene intron region 1 CpG Island, aiming to investigate its correlations with the incidence of EOPD.
Materials and Methods
Subjects
According to the diagnostic criteria of BrainBank (UK) [9], 40 inpatients and outpatients (mean age 41.23 ± 5.27 y, 20 males and 20 females) treated in the department of neurology, The Affiliated Tumor Hospital of Xinjiang Medical University, from January 2010 to August 2013 and 40 healthy volunteers (mean age 44.13 ± 4.89 y, 20 males and 20 females) were included in this study. This study was conducted in accordance with the declaration of Helsinki. This study was conducted with approval from the Ethics Committee of Xinjiang Medical University. Written informed consent was obtained from all participants.
DNA extraction
The QIAamp DNA Mini Kit (QIAGEN, Dusseldorf, Germany) kit was used to extract DNA from the blood samples, followed by quantification using a spectrophotometer; 100 ng of DNA was then performed electrophoresis using 0.8% agarose gel for the quality inspection, and the qualified DNA was then adjusted to 75 ng/µl, transferred into 384-well plates, and stored at -20°C for future use.
Methylation analysis
After treated with sulfite, the DNA sample was performed PCR amplification, alkaline phosphatase treatment, in vitro transcription, and RNase digestion as well as purification; the products were then loaded into the 384-spot SpectroCHIP (SEQUENOM, Santiago California, USA) and automatically detected and analysed using Mass ARRAY Compact System (SEQUENOM); the EpiTYPER software (SEQUENOM) was then used to analyse and output the results.
Statistical analysis
This study was one case-control study, so the experimental data were analysed using SPSS18.0 statistical package. The measurement data of the two groups were firstly tested the homogeneity of variance, and the data that met the criteria were then performed the t test, otherwise, the rank sum test was used for the comparison.
Results
Sequence of SNCA gene intron 1
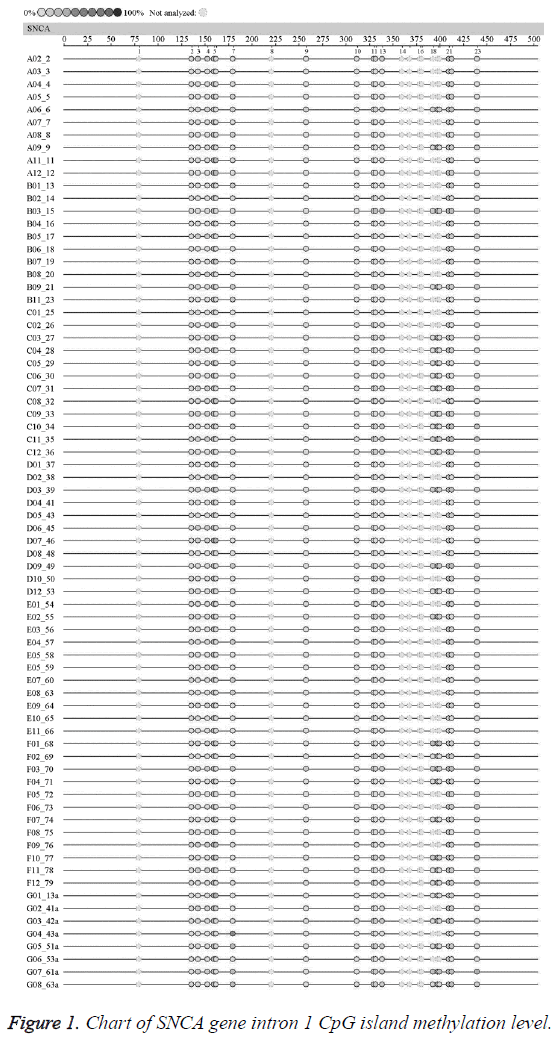
In order to further investigate the methylation levels of SNCA gene intron l CpG Island in the EOPD patients and the normal control, this study firstly analysed the fragments within the -926~-476 loci in SNCA intron region 1. Previous studies reported that CG within SNCA intron region 1 926-476 sites were found hypomethylated in the brain tissues of sporadic PD patients, and the hypomethylation in this region might be associated with the abnormal upregulation of SNCA mRNA, which might possibly participate in regulating the SNCA gene expression [6]. This region is located within l kb of the upstream of the transcription start point, containing 23 CpG Island loci. The location and sequence of SNCA gene intron 1: SNCA intron 1 (-926~-476) is located within l kb of the upstream of the transcription start site (ATG); the -926~-476 region contains 23 CpG island loci (marked in red), and the results were shown in Table 1.
| SNCA methylation list |
|---|
| 926- GgagcctaaggaaagagacttgacctggctttcgtcctgcttctgatattcccttctccacaagggctgagagattaggctgCttctccgggatccgcttttccccgggaaacgcgaggatgctccatggagcgtgagcatccaacttttctctcacat aaaatctgtctgcccgctCtcttggtttttctctgtaaagtaagcaagctgcgtttggcaaataatgaaatggaagtGcaaggaggccaagtcaacaggtggtaacgggttaacaagtgctggcgcggggtccgctAgggtggaggctgagaac gccccctcgggtggctggcgcggggttggagacggcccgcgAgtgtgagcggcgcctgctcagggtagatagctgagggcgggggtggatgttggatgttGgatggattagaaccatcacacttgggcctgctgtttg-476 |
Table 1: SNCA methylation list.
Methylation level chart of SNCA gene intron 1
The average methylation levels of all the 23 CpG island loci in SNCA gene intron region l in the EOPD patients and the normal control were shown in Figure 1. The undetected sites were colorless; according to the methylation color scale in the upper left corner, the deeper color indicated the higher methylation degree of this CpG Island. The results suggested no significant difference in the methylation level chart of each CpG island in SNCA gene intron 1 between the EOPD group and the control group.
Average SNCA intron region 1 methylation level
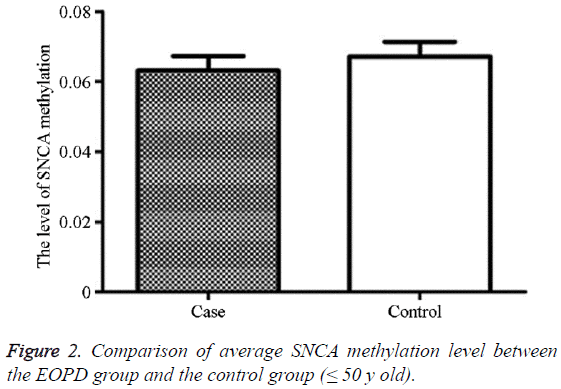
The average methylation level in the EOPD patients was 6.31% (2.56, 14.56), and that in the control group was 6.71% (1.89, 13.00), showing no statistically significant difference (P=0.998) (Table 2 and Figure 2).
| Group | N | Average methylation level of SNCA (%) | P |
|---|---|---|---|
| EOPD | 40 | 6.31 (2.56, 14.56) | 0.998 |
| Control | 40 | 6.71 (1.89, 13.00) |
Table 2: Comparison of average SNCA intron region 1methylation level between the EOPD group and the control group (≤ 50 y old).
Average SNCA intron region 1 methylation level in male
The average methylation level in the male EOPD patients was 6.52% (2.56, 14.56), and that in the control group was 6.28% (1.89, 12.44), showing no statistically significant difference (P=0.225) (Table 3).
| Group | N | Average methylation level of SNCA (%) | P |
|---|---|---|---|
| Male EOPD | 21 | 6.52 (2.56, 14.56) | 0.225 |
| Male control | 21 | 6.28 (1.89, 12.44) |
Table 3: Comparison of average SNCA intron region 1 methylation level in male between the EOPD group and the control group (≤ 50 y old).
Average SNCA intron region 1 methylation level in female
The average methylation level in the female EOPD patients was 6.38% (2.78, 9.11), and that in the control group was 7.19% (4.33, 13.00), showing no statistically significant difference (P=0.296) (Table 4).
| Group | N | Average methylation level of SNCA (%) | P |
|---|---|---|---|
| Female EOPD | 19 | 6.38 (2.78, 9.11) | 0.296 |
| Female control | 19 | 7.19 (4.33, 13.00) |
Table 4: Comparison of average SNCA intron region 1 methylation level in female between the EOPD group and the control group (≤ 50 y old).
Average SNCA intron region 1 methylation level in Han
The average methylation level in Han EOPD patients was 5.51% (5.00, 13.00), and that in the control group was 6.54% (4.44, 9.44), showing no statistically significant difference (P=0.520) (Table 5).
| Group | N | Average methylation level of SNCA (%) | P |
|---|---|---|---|
| EOPD patients in Han | 21 | 5.51 (5.00, 13.00) | 0.52 |
| Control in Han | 21 | 6.54 (4.44, 9.44) |
Table 5: Comparison of average SNCA intron region 1 methylation level between the EOPD group and the control people in Han (≤ 50 y old).
Average SNCA intron region 1 methylation level in Uighur
The average methylation level in Uighur EOPD patients was 7.19% (1.89, 12.11), and that in the control group was 6.38% (2.56, 14.56), showing no statistically significant difference (P=0.948) (Table 6).
| Group | N | Average methylation level of SNCA (%) | P |
|---|---|---|---|
| EOPD patients in Uighur | 19 | 7.19 (1.89, 12.11) | 0.948 |
| Control in Uighur | 19 | 6.38 (2.56, 14.56) |
Table 6: Comparison of average SNCA intron region 1 methylation level between the EOPD group and the control people in Uighur (≤ 50 y old).
Average SNCA intron region 1 methylation level between the male and female
The average methylation level in the male EOPD patients was 7.19% (4.33, 13.00), and that in the female EOPD patients was 6.28% (1.89, 12.44), showing no statistically significant difference (P=0.948) (Table 7).
| Group | N | Average methylation level of SNCA (%) | P |
|---|---|---|---|
| Male EOPD patients | 21 | 7.19 (4.33, 13.00) | 0.051 |
| Female EOPD patients | 19 | 6.28 (1.89, 12.44) |
Table 7: Comparison of average SNCA intron region 1 methylation level between the male and female EOPD patients.
Average SNCA intron region 1 methylation level between the Han and the Uighur
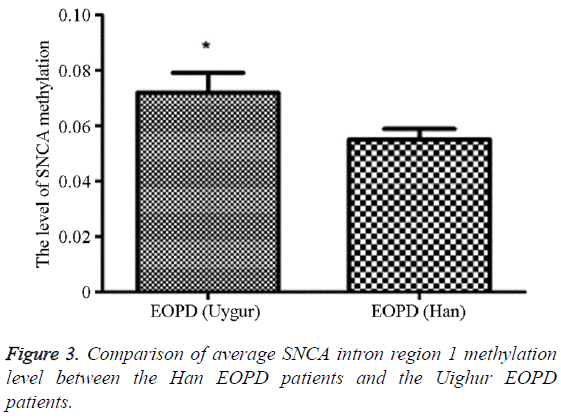
The methylation average level in the Uighur EOPD patients was 5.51% (5.00, 13.00), and that in the Han EOPD patients was 7.19% (1.89, 12.11), which was significantly higher than the average level in the Uighur EOPD patients, and the difference was statistically significant (P=0.020, OR (95% CI)=0.81 (0.03-3.35)) (Table 8 and Figure 3).
| Group | N | Average methylation level of SNCA (%) | P | OR (95% CI) |
|---|---|---|---|---|
| Uighur EOPD patients | 19 | 7.19 (1.89, 12.11) | 0.02 | 0.81 (0.03-3.35) |
| Han EOPD patients | 21 | 5.51 (5.00, 13.00) |
Table 8: Comparison of average SNCA intron region 1 methylation level between the Han EOPD patients and the Uighur EOPD patients.
Discussion
The reason of sporadic PD is still unclear, and currently it is generally considered as the result of the interactions of environmental factors and genetic susceptibility [10]. New evidence suggests that chronic environmental exposure could alter gene expressions through certain epigenetic mechanisms, and it might be a key factor for the delayed neurodegenerative diseases. Studies found the number of the DNA methylation sites increased with aging, and these changes might contribute to the aging process, as well as an important risk factor in the pathogenesis of PD, or they might be the stress responses appearing in the aging process. The whole genome research that determined the candidate gene methylation revealed the changes of the methylation status in the proximal DNA CpG island loci of the RAB7L1, GPNMB, and STX1B candidate genes. Maeda et al. [11] found the existence of subtelomeric DNA abnormal methylation in the peripheral blood cells of PD patients; therefore, it was speculated that in PD patients, oxidative stress caused subtelomeric DNA methylation disorder was closely associated with the degeneration of nerve cells. Studies had reported the DNA methylation abnormalities in neurological diseases. For example, the postmortem brain tissues of patients with AD [12,13], PD, Multiple Sclerosis (MS) [14,15] are due to abnormal disease-related gene DNA methylation. These results suggested that the DNA methylation status in the peripheral blood cells could become a biomarker of a number of neurological diseases [16,17].
Presently, certain studies had revealed that the diploid and triploid of SNCA gene and the missense mutation of A53T, A30P, and E46K were the pathological basis of the autosomal dominant forms of PD and Lewy Bodies (LB) [18-20]. Although the connections between the DNA methylation and the PD pathogenesis is still unclear, the methylation of SNCA gene might possibly cause structural changes or overexpress certain proteins, thus resulting in the protein aggregation, or involve in the pathogenesis of PD by gene expression disorders. The methylation of SNCA intron 1 had been confirmed to be able to reduce the SNCA transcription, and it was also found that in the cerebral substantia nigra of sporadic PD patients, the methylation level of SNCA intron 1 was reduced, which led to the increased expression of SNCA gene [6]. Therefore, it could be considered that the reduced SNCA gene methylation would increase the expression of SNCA gene, thus resulting in an increase in the production of α- synuclein and the following onset of PD.
Previous studies also suggested that different methylation levels of SNCA intron region 1 might be associated with the occurrence of EOPD [7], so our group further investigated the correlations of this methylation with the EOPD risks in Uighur and Han populations; the results showed that the average methylation level in EOPD patients was 6.31%, while that in the control group was 6.71%, and the difference was no statistically significant (P=0.998), consistent with Richter et al. [8] while different from Jowaed et al. [6], whose study found that the methylation level of SNCA intron 1 in the cerebral substantia nigra of PD patients was significantly reduced, but the former reported no significant difference in the methylation status of this fragment in the peripheral WBC between the PD patients and normal controls using the pyrophosphate sequencing technique, so the links between the SNCA intron 1 methylation level and the incidence of PD could not be identified. This might also be related with the confounding factor controlling in the study subjects; in addition, it is extremely difficult to obtain brain tissue samples from living people, so the above reports could not be confirmed by other studies. We also cannot rule out the impact of environmental factors on methylation and multi-center large sample validation was needed to verification. In order to further investigate the associations between the methylation level in this region and the PD susceptibilities in different ethnic groups, this study performed stratified analysis and comparison towards different nationalities, and found that the average methylation level in the Uighur EOPD group was 7,19% (1.89, 12.11), while that in the Han EOPD group was 5.51% (5.00, 13.00), statistically significantly lower than the methylation level in the Uighur EOPD group (P=0.020, OR (95% CI)=0.81 (0.03-3.35)), and it was considered to be caused by the fact that Xinjiang is located in Central Asia, so that the Uighur has different genetic backgrounds from Han, so the geographical and racial differences might result in the different results. The results of this study suggested that the hypomethylation of SNCA gene in PBMCs of the Uighur nationality might be a characteristic of sporadic Uighur PD patients. The results of this study indicated that the methylation of SNCA gene was not connected with the occurrence of PD in the Uighur population, especially those younger than 50 y old; however, whether the SNCA gene methylation in PBMCs of the sporadic Uighur and Han PD patients was related with PD still needed to be further investigated with larger sample sizes.
Funding
This study was supported by grants from Xinjiang Uygur Autonomous Region Science and Technology Supporting Project in Xinjiang (No. 201591160).
Conflict of Interest
All authors have no conflict of interest regarding this paper.
References
- Arenas E. Towards stem cell replacement therapies for Parkinsons disease. Biochem Biophys Res Commun 2010; 396: 152-156.
- Singh PK, Kotia V, Ghosh D, Mohite GM, Kumar A, Maji SK. Curcumin modulates a-synuclein aggregation and toxicity. ACS Chem Neurosci 2012; 4: 393-407.
- Federico A, Cardaioli E, Da Pozzo P, Formichi P, Gallus GN. Mitochondria, oxidative stress and neurodegeneration. J Neurol Sci 2012; 322: 254-262.
- Hauser DN, Hastings TG. Mitochondrial dysfunction and oxidative stress in Parkinsons disease and monogenic Parkinsonism. Neurobiol Dis 2013; 51: 35-42.
- Masliah E, Dumaop W, Galasko D, Desplats P. Distinctive patterns of DNA methylation associated with Parkinson disease: identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013; 8: 1030-1038.
- Jowaed A, Schmitt I, Kaut O, Wüllner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinsons disease patients’ brains. J Neurosci 2010; 30: 6355-6359.
- Matsumoto L, Takuma H, Tamaoka A, Kurisaki H, Date H, Tsuji S, Iwata A. CpG demethylation enhances alpha-aynuclein expression and affects the pathogenesis of Pakinsons disease. PLoS One 2010; 5: 15522.
- Richter-Landsberg C, Gorath M, Trojanowski JQ, Lee VM. alpha-synuclein is developmentally expressed in cultured rat brain oligodendrocytes. J Neurosci Res 2000; 62: 9-14.
- Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinsons disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 1992; 55: 181-184.
- de Lau LM, Breteler MM. Epidemiology of Parkinsons disease. Lancet Neurol 2006; 5: 525-535.
- Maeda T, Guan JZ, Oyama J, Higuchi Y, Makino N. Aging-associated alteration of subtelomeric methylation in Parkinsons disease. J Gerontol A Biol Sci Med Sci 2009; 64: 949-955.
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD. Epigenetic changes in Alzheimers disease: decrements in DNA methylation. Neurobiol Aging 2010; 31: 2025-2037.
- West RL, Lee JM, Maroun LE. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimers disease patient. J Mol Neurosci 1995; 6: 141-146.
- Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res 2007; 85: 2006-2016.
- Casaccia-Bonnefil P, Pandozy G, Mastronardi F. Evaluating epigenetic landmarks in the brain of multiple sclerosis patients: a contribution to the current debate on disease pathogenesis. Prog Neurobiol 2008; 86: 368-378.
- Ai S, Shen L, Guo J, Feng X, Tang B. DNA methylation as a biomarker for neuropsychiatric diseases. Int J Neurosci 2012; 122: 165-176.
- Tan YY, Wu L, Zhao ZB, Wang Y, Xiao Q, Liu J, Wang G, Ma JF, Chen SD. Methylation of a-synuclein and leucine-rich repeat kinase 2 in leukocyte DNA of Parkinsons disease patients. Parkinsonism Relat Disord 2014; 20: 308-313.
- Thomas B, Beal MF. Molecular insights into Parkinsons disease. F1000 Med Rep 2011; 3: 7.
- Tateno F, Sakakibara R, Kawai T, Kishi M, Murano T. Alpha-synuclein in the cerebrospinal fluid differentiates synucleinopathies (Parkinson Disease, dementia with Lewy bodies, multiple system atrophy) from Alzheimer disease. Alzheimer Dis Assoc Disord 2012; 26: 213-216.
- Tijero B, Gomez-Esteban JC, Llorens V, Lezcano E, Gonzalez-Fernandez MC, de Pancorbo MM, Ruiz-Martinez J, Cembellin JC, Zarranz JJ. Cardiac sympathetic denervation precedes nigrostriatal loss in the E46K mutation of the a-synuclein gene (SNCA). Clin Auton Res 2010; 20: 267-269.