Research Article - Microbiology: Current Research (2018) Volume 2, Issue 2
Bacteriological and antibiogram of AmpC producing Enterobacteriaceae isolated from abattoir.
Ejikeugwu C1*, Nworie O.2, Agah M.V.1, Oguejiofor B.1, Ovia K.3, Nworie C.O1, Iwunze A.C.1, Nwambeke A.1 and Edeh C1
1Department of Applied Microbiology, Faculty of Science, Ebonyi State University, Abakaliki, P.M.B 053, Ebonyi State, Nigeria
2Department of Biology, Microbiology and Biotechnology, Faculty of Science, Federal University, Ndufu-Alike, Ikwo, Nigeria
3Department of Biological Sciences, Evangel University, Akaeze, Ebonyi State, Nigeria
- *Corresponding Author:
- Ejikeugwu Chika
Faculty of Science
Department of Applied Microbiology
Ebonyi State University, Nigeria
Tel: +2348097684562
Email: ejikeugwu_chika@yahoo.com
Accepted date: January 29, 2018
Citation: Ejikeugwu C, Nworie O, Agah MV, et al.. Bacteriological and antibiogram of AmpC producing Enterobacteriaceae isolated from abattoir. Microbiol Curr Res. 2018;2(2):30-34.
Abstract
Owing to the continued threat of antimicrobial resistance, it is critical that farmers and clinicians take the necessary steps to use antibiotics rationally in their respective practices, and only opt for it when all other options must have been exhausted. AmpC enzymes mediate bacteria resistance to the cephamycins such as cefotetan and cefoxitin which are important antibiotics used clinically to treat and manage bacterial related infections. This study phenotypically evaluated the prevalence and antibiogram of Enterobacteriaceae that produced AmpC enzymes. Fifty (50) anal swab samples from the anal region of cows in an abattoir in Abakaliki, Nigeria were bacteriologically analyzed. The isolation and identification of bacteria isolates were carried out using standard microbiology techniques. Antibiogram was evaluated using disk diffusion technique while AmpC enzyme production was detected using the ceftazidime imipenem antagonism test. Our results show that a total of 7 (14%) E. coli and 12 (24%) isolates of Klebsiella species was isolated. The antibiogram results showed that the isolated Klebsiella species and E. coli isolates exhibited reduced susceptibility to cloxacillin (100%), ertapenem (83.3%), cefoxitin (66.7%) and ceftazidime (58.3%) for Klebsiella species; and cefoxitin (71.4%), ertapenem (28.6%) and ceftazidime (28.6%) for isolates of E. coli. AmpC enzyme production was detected in 5 (71%) isolates of E. coli and 9 (75%) isolates of Klebsiella species. The AmpC producing E. coli and Klebsiella species were multiply resistant to over 4 antibiotics found in the class: fluoroquinolones, carbapenems, aminoglycosides and penicillins. Antibiotic usage in animal husbandry allows drug resistant bacteria such as those that produce AmpC enzymes to evolve and spread. This has implication for the general public since AmpC producing bacteria are notably resistant to 2nd generation cephalosporins which are used clinically for the treatment of serious bacterial infections. Continuous monitoring of the antibiotic resistance profile in abattoir and poultry isolates is recommended as a panacea to contain the problem of antibiotic resistance in the non-hospital environment.
Keywords
AmpC enzymes, Escherichia coli, Klebsiella species, Antibiotic Resistance
Introduction
A remarkable therapeutic success heralded the introduction of antibiotics into clinical medicine as a panacea to treating and containing the nefarious activities of pathogenic microbes. Irrespective of the help of antibiotics in reducing the rate of morbidity and mortality due to infectious disease agents especially bacteria, some bacterial pathogens have evolved mechanisms with which to become resistant to the antimicrobial onslaught of antimicrobial agents in their environment.This phenomenon has allowed some multidrug resistant bacterial pathogens including those that produce AmpC enzymes to emerge and spread almost uncontrollably. Antibiotics have been used extensively in veterinary medicine and in agriculture; and their usage in livestock feeds at sub-therapeutic doses to promote growth and increase feed efficiency has contributed to antimicrobial resistance cases [1-3]. AmpC β-lactamases are produced by many Enterobacteriaceae strains, and they mediate bacterial resistance to cefotetan and cefoxitin [4-6]. They are carried on the genetic elements of many Enterobacteriaceae and some other bacteria such as Pseudomonas and Acinetobacter where they cause resistance to cephamycins and other betalactam agents [7-9]. The non-medical uses of antimicrobial agents allow bacteria to develop resistance through selective pressure. Previous reports on the prevalence and spread of drug resistant bacteria in the community give impetus to the irrational use of antimicrobial agents for non-clinical purposes [10,11]. The prevalence of infections with multidrug resistant Enterobacteriaceae has steadily increased over the years; and this has impacted negatively on the health of the general public [11-14]. Enterobacteriaceae producing AmpC beta-lactamases have become a major therapeutic challenge as well, since this type of resistance mechanism mediate bacterial resistance to the cephamycins. The detection and reporting of AmpC-producing Gram negative bacteria is therefore of significant clinical relevance since AmpC producers may appear susceptible to expanded-spectrum cephalosporins when initially tested [15]. In this study, the prevalence of AmpC-producing Klebsiella species and E. coli isolates were phenotypically investigated.
Collection and processing of samples
The samples (n=50) used for this study were collected from the anal region of cows from a local abattoir in Abakaliki metropolis, Ebonyi State, Nigeria. The samples were collected using sterile swab sticks soaked in normal saline. The collected samples was returned to their respective containers and labeled. All samples were bacteriologically analyzed within one hour of collection at the Microbiology Laboratory Unit of Ebonyi State University, Abakaliki. Each of the collected samples was inserted into 5 ml of freshly prepared nutrient broth (Oxoid, UK), and incubated at 30°C overnight. Bacterial growth was identified by the presence of turbidity or cloudiness in the broth culture after incubation [16].
Culture and characterization
The turbid solution from the overnight broth culture was inoculated aseptically on MacConkey agar (MAC) and eosin methylene blue (EMB) agar (Oxoid, UK) plates, and incubated at 30°C overnight. Suspect colonies of Escherichia coli and Klebsiella species were subcultured onto freshly prepared MAC and EMB agar plates for the isolation of pure cultures of E. coli and Klebsiella species. E. coli and Klebsiella species isolates were identified based on their colonial, biochemical, microscopical and morphological characteristics [16].
Antimicrobial susceptibility testing
This was performed using amikacin (AK, 30 µg), cefoxitin (FOX, 30 µg), cloxacillin (OB, 10 µg), ceftazidime (CAZ, 30 µg), ofloxacin (OFX, 5 µg), ertapenem (ETP, 10 µg) and imipenem (IPM, 10 µg) (Oxoid, UK). The guidelines of the Clinical and Laboratory Standard Institute (CLSI) were used for antimicrobial susceptibility testing. The antimicrobial susceptibility testing was performed as was previously described [17,18].
Screening test
Bacterial strains that produce AmpC beta-lactamase enzymes are resistant to the cephamycins but susceptible to the fourth generation cephalosporin, cefepime [9,17]. The susceptibility of the test isolates to cefoxitin disk (30 µg) was used as the primary screening test to screen all the isolates for possible production of AmpC enzymes. All bacterial isolates were each subjected to the antimicrobial activity of cefoxitin disk (30 µg) on an aseptically streaked MH agar plates, and these were incubated at 30°C for 18-24 h. The production of AmpC enzyme was suspected in those test isolates that showed resistance to cefoxitin disk based on the CLSI breakpoints [17,18]. Test isolates showing inhibition zone diameter (IZD)<18 mm were suspected for the production of AmpC beta-lactamase enzyme.
Confirmatory test for AmpC enzyme production
Ceftazidime-imipenem antagonism test (CIAT) was used to confirm AmpC enzyme production in the bacterial isolates. This was performed using ceftazidime (30 µg), cefoxitin (30 µg) and imipenem (10 µg) (Oxoid, UK). Ceftazidime disc and imipenem disk were placed at a distance of 20 mm apart on MH agar plate previously inoculated with a suspension of the test bacteria (adjusted to 0.5 McFarland turbidity standards). Cefoxitin disk (30 µg) was also placed at a distance of 20 mm from the ceftazidime disk for comparison. Incubation was at 30°C overnight. AmpC beta-lactamase production was inferred by antagonism indicated by a visible reduction in the inhibition zone around the ceftazidime disk adjacent to the imipenem or cefoxitin disk [5].
Calculation of Multiple Antibiotic Resistance Index (MARI)
This was calculated to determine the multiple antibiotic resistance profile of the isolated Klebsiella species and E. coli isolates that were positive for AmpC enzyme production. This was done according to the method of Akinjogunla and Enabulele [19]. MARI was calculated using the formula: MARI=a/b;where ‘a’ represents the number of antibiotics which the resistant bacteria was resistant to; and ‘b’ represents the total number of antibiotics to which the resistant bacteria has been evaluated for.
Results
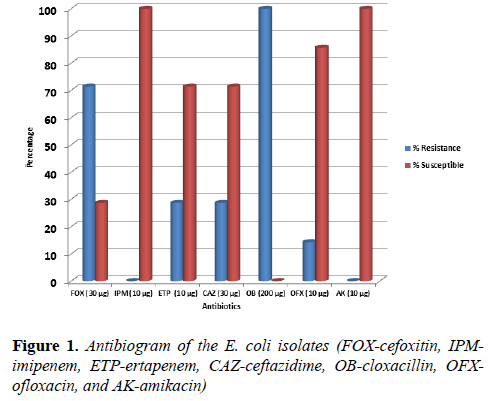
The distribution of the number of bacterial isolates recovered from the anal swab samples is shown in Table 1. A total of 7 isolates of E. coli and 12 isolates of Klebsiella species were isolated from the samples. The E. coli isolates were positive for indole production and methyl red test, while the Klebsiella species isolates were positive for citrate and urease production. Figure 1 shows the antibiogram of the isolated E. coli isolates to selected test antibiotics.
| Bacteria | Number (%) | Source | Gram reaction |
|---|---|---|---|
| Escherichia coli | 7 (14) | Abattoir | Negative |
| Klebsiella species | 12 (24) | Abattoir | Negative |
Table 1: Frequency of isolation of klebsiella species and E. coli isolates
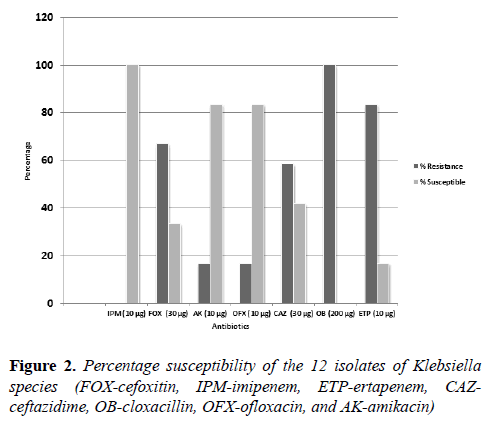
The E. coli isolates were resistant to ofloxacin (14.3 %), ceftazidime (28.6 %), ertapenem (28.6 %) and cefoxitin (71.4 %). They were also resistant to cloxacillin (100 %). None of the E. coli isolates were resistant to imipenem and amikacin, as they were found to be completely susceptible to these antibiotics found in the carbapenem and aminoglycoside family respectively (Figure 1). The antibiogram of the 12 isolates of Klebsiella species is shown in Figure 2.
Isolates of Klebsiella species were found to be resistant to cefoxitin (66.7%), cloxacillin (100%), ertapenem (83.3%) and ceftazidime (58.3%). The Klebsiella species isolates were all susceptible to imipenem as seen in the E. coli isolates. The isolates of Klebsiella species also showed decreased susceptibility to amikacin (16.7%) and ofloxacin (16.7%), which are antibiotics that belong to the aminoglycoside and fluoroquinolone classes (Figure 2). The prevalence of AmpC producing Enterobacteriaceae of E. coli and Klebsiella species in this study is shown in Table 2.
| Bacteria (n) | AmpC positive n (%) | AmpC negative n (%) |
|---|---|---|
| Escherichia coli (7) | 5 (71) | 2 (29) |
| Klebsiella species (12) | 9 (75) | 3 (25) |
Table 2: Occurrence of AmpC producing bacteria
Of the 7 E. coli isolates screened and phenotypically confirmed for MBL production, only 5 (71%) isolates of E. coli were positive for AmpC enzyme production by the ceftazidime imipenem antagonism test (CIAT). The production of AmpC enzymes was confirmed in 9 (75%) isolates of Klebsiella species out of the 12 isolates of Klebsiella species screened for the production of AmpC enzymes in this study (Table 2). Table 3 shows the results of the multiple antibiotic resistance nature of the AmpC positive E. coli and Klebsiella species. The AmpC positive E. coli isolates were multiply resistant to 4 antibiotics on average out of the 7 antibiotics used in this study. On average however, the isolates of Klebsiella species were multiply resistant to 5 antibiotics out of the 7 antibiotics used in this study (Table 3).
| Isolate number | MARI |
|---|---|
| K2 | 0.1 |
| K3 | 0.6 |
| K4 | 0.6 |
| K9 | 0.4 |
| K10 | 0.7 |
| K11 | 0.6 |
| K13 | 0.4 |
| K17 | 0.6 |
| K23 | 0.6 |
| E2 | 0.4 |
| E7 | 0.6 |
| E5 | 0.3 |
| E3 | 0.3 |
| E4 | 0.4 |
Table 3: Multiple antibiotic resistance index of AmpC producing E. coli and klebsiella species
Discussion
The distribution of antibiotic resistant bacteria and their genetic factors in the non-hospital environment is attributable to the use of antibiotics especially irrationally. Antibiotics are usually overused in humans and animals especially for prophylactic measures and as growth promoting agents. These factors have significantly contributed to the evolving nature of drug resistant pathogens including those that produce AmpC enzymes as well as their unfavourable spread in both the hospital and non-hospital environment. In this study, we evaluated by phenotypic techniques the antibiogram and prevalence of AmpC producing E. coli and Klebsiella species isolates. E. coli and Klebsiella species were bacteriologically recovered from the anal swab samples at the rates of 14% and 24% respectively, which is parallel to our earliest reports conducted in 2016 and 2017 [5,17]. These bacteria are members of the Enterobacteriaceae family and part of the human normal flora. However, antibiotic resistance in bacteria from the community could contribute to some community-acquired infections, and thus render some available antibiotics inefficacious when used for therapy. These bacteria can also be transmitted through contact with already infected animals. The isolated E. coli in our study was resistant to some cephalosporins and carbapenems including ceftazidime (28.6%), ertapenem (28.6%) and cefoxitin (71.4%). Resistance to the fluoroquinolone, ofloxacin (14.3%) was also recorded amongst the E. coli isolates. Most notably was the observation that none of the E. coli isolates was resistant to imipenem and amikacin, which are members of the carbapenem and aminoglycoside class of antibiotics. And they are both used to treat infections caused by this organism. The E. coli isolates were susceptible to cloxacillin (100%) which is a beta-lactam drug that is not routinely used for treatment of E. coli infections but may be included in some multidrug resistant infections in which this organism has been implicated [20-22]. Higher resistance rates were also observed among the isolates of Klebsiella species used in this study. Nearly 83.3% and 66.7% of the isolates of Klebsiella species were resistant to ertapenem and cefoxitin respectively. These are antibiotics used to treat Klebsiella species infections [21,22]. The isolates of Klebsiella species also showed reduced susceptibility to ceftazidime (58.3%), cloxacillin (100%), amikacin (16.7%) and ofloxacin (16.7%). All the isolates of Klebsiella species in this study were completely susceptible to imipenem, a carbapenem that is used to treat serious infections caused by Klebsiella species [21,23]. The antibiograms of in various other studies showed resistance of these organisms towards most of the antibiotics [1,10,24,25]. AmpC producing bacteria in this study was detected at 71% and 75% for isolates of E. coli and Klebsiella species respectively. Notably, only 5 (71%) isolates of E. coli produced AmpC enzymes out of the 7 isolates phenotypically screened for the enzyme while 9 (75%) isolates of Klebsiella species produced AmpC enzymes out of the 12 isolates phenotypically screened for the enzyme production. This is similar to an earlier report by Tan et al. [12] and Adler et al. [26] in which AmpC enzymes were significantly detected in the E. coli and Klebsiella species in Asia and Europe respectively. In one of our earlier reports in 2016, we had reported the occurrence of AmpC producing bacteria in the Enterobacteriaceae family in which AmpC enzymes was phenotypically detected in isolates of Klebsiella species from abattoir samples in Abakaliki, Nigeria. The Klebsiella species and E. coli isolates positive for AmpC enzyme production were multiply resistant to more than 4 antibiotics out of 7 antibiotics. This shows that the AmpC producing E. coli and Klebsiella species isolated in this study are multidrug resistant. Conclusively, this study show that isolates of E. coli and Klebsiella species from abattoir are multidrug resistant. They also produce AmpC enzymes which allow them to be resistant to the 2nd generation cephalosporins which are clinically used to manage and treat serious bacterial infections. This present study is significant and gives impetus to the growing frequency of antibiotic resistance in the non-hospital environment such as abattoir. It also gives credence to the possible abuse and irrational use of antibiotics in animal husbandry and for other non-clinical purposes. These practices allow antibiotic resistant bacteria to emerge and spread in the community; and such organisms could be implicated in a handful of community-acquired infections. Adequate surveillance and detection measures are needed to decipher and possibly contain the evolving nature and spread of resistant bacteria in this environment, particularly those that produce AmpC enzymes.
References
- Manus M, Stockwell PS, Sundin VO, et al. Antibiotic use in plant agriculture. Annu Rev of Phytopathol. 2002;40:443-65.
- Geser N, Stephan R, Zbinden R, et al. Fecal carriage of extended-spectrum beta- lactamase (ESBL) producing Enterobacteriaceae in swine and cattle at slaughter in Switzerland. J Food Protect. 2011;74:446-9.
- Paterson L, Hujer M, Yeiser B, et al. Extended-spectrum beta- lactamases. Dominance and widespread prevalence of SHV- and CTX-M- type beta - lactamases. Antimicrob Agents Chemother. 2003;47:3554-60.
- Hemalatha V, Padma M, Sekar U, et al. Detection of AmpC beta lactamase production in Escherichia coli and Klebsiella by an inhibitor based methods. Indian J Med Res. 2007;126:220-3.
- Ejikeugwu C, Duru C, Eluu S, et al. Isolation and Phenotypic Detection of Metallo-Beta Lactamase (MBL)-Producing Klebsiella Species from Cow Anal Swabs. Global J of Pharma Sci. 2017;2:1-5.
- Papanicolaou GA, Medeiros AA, Jacoby GA. Novel plasmid mediated β-lactamases (MIR-1) conferring resistance to oxyimino and α methoxy β-lactams in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1990;34:2200–9.
- Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–82.
- Caroff N, Espaze E, Gautreau D, et al. Analysis of the effects of 42 and 32 AmpC promoter mutations in clinical isolates of Escherichia coli-hyper-producing AmpC. J Antimicrob Chemother. 2000;45:783–8.
- Jacoby GA, Munoz-Price LS. Mechanisms of Disease: The New β–Lactamases. N Engl J Med. 2005;352:380-91.
- Hyun-Ho S, Seung-Hak C. Prevalence of antimicrobial resistance in Escherichia coli strains isolated from fishery workers. Osong Public Health Res Perspect. 2013;4:72-5.
- Pfaller MA, Segreti J. Overview of the epidemiological profile and laboratory detection of extended-spectrum beta-lactamases. Clin Infect Dis. 2006;42:153–63.
- Tan T, Ng Y, Teo SY, et al. Detection of plasmid-mediated AmpC in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis. J Clin Pathol. 2009;61:642–4.
- Siu LK, Lu PL, Chen JY, et al. High-level expression of ampC beta-lactamase due to insertion of nucleotides between 10 and 35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrum-cephalosporin treatment. Antimicrob Agents Chemother. 2003;47:2138–44.
- Perez-Perez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–62.
- Thomson KS. Controversies about extended-spectrum and AmpC beta-lactamases. Emerg Infect Dis. 2001;7:333–6.
- Cheesbrough M. District laboratory practice in tropical countries. Cambridge University Press, UK. 2006;178-87.
- Ejikeugwu C, Iroha I, Ugwu M, et al. Phenotypic detection of AmpC enzymes and antimicrobial susceptibility of Klebsiella spp. isolated from abattoir. Int J of Microbiol and Biotechnol Res. 2016;4:117-21.
- Wayne PA. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing: Approved standard (M100-S20) USA. 2011
- Akinjogunla OJ, Enabulele IO. Virulence factors, plasmid profiling and curing analysis of multidrug resistant Staphylococcus aureus and coagulase negative Staphylococcus spp. isolated from patients with Acute Otitis Media. J of Am Sci. 2010;6:1022-33.
- Ryan KJ, Ray CG. Sherris Medical Microbiology (4th ed.). McGraw Hill Publishers,USA. 2004;370.
- Brooks GF, Butel JS, Morse SA, et al. Medical Microbiology(23rd ed). McGraw Hill Publishers, USA. 2004;248-60.
- Zanetti G, Bally F, Greub G. Cefepime versus imipenem-cilastatin for treatment of nosocomial pneumonia in intensive care unit patients (a multicenter, evaluator-blind, prospective, randomized study). Antimicrob Agents Chemother. 2003;47:3442–7.
- Walsh TR, Toleman MA, Poirel L, et al. Metallo β – Lactamases: The Quiet Before the Storm. Clin Microbio Rev. 2005;18:306-25.
- Livermore DM, James D, Reacher M, et al. Trends in fluoroquinolone (ciprofloxacin) resistance in Enterobacteriaceae from bacteremias. Emergence of Infect Dis. 2002;8:473-8.
- Miftode E, Dorneanu O, Leca D, et al. Antimicrobial resistance profile of E. coli and Klebsiella spp. from urine in the Infectious Diseases Hospital Iasi. Rev Med Chir Soc Med Nat Iasi. 2008;113:478-82.
- Adler H, Fenner L, Walter P, et al. Plasmid-mediated AmpC β-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: prevalence at a Swiss university hospital and occurrence of the different molecular types in Switzerland. J Antimicrob Chemother. 2008;61:457-8.