Research Article - Biomedical Research (2017) Volume 28, Issue 2
A meta-analysis of the TFPI2 hyper-methylation frequency and colorectal cancer risk
Niancai Jing1#, Tao Huang2#, Yi Lu1, Hongyu Xiao1, Huanyu Guo1, Zhuo Chen1, Yue Zhang1*1Tumor Hospital of Jilin Province, Changchun, Jilin, P.R.China
2Affiliated Hospital of Changchun University of Traditional Chinese Medicine, Changchun, Jilin, P.R.China
#These authors contributed equally to this work and should be considered co-first authors
- *Corresponding Author:
- Yue Zhang
Tumor Hospital of Jilin Province
Changchun, Jilin, P.R China
E-mail: zhangyue160712@163.com
Accepted date: July 26, 2016
Abstract
Tissue Factor Pathway Inhibitor 2 (TFPI2) is a serine protease inhibitor; it plays an important role in the degradation and remodeling of Extracellular Matrix (ECM). The degradation and reshaping of ECM is the basic steps of tumour growth and metastasis, angiogenesis, cell migration, and normal physiological processes. Therefore, TFPI2 plays a key in cancer, while it often silence by its promoter methylation. This study aimed to conduct a meta-analysis to access the relationship between TFPI2 promoter hyper-methylation and Colorectal Cancer (CRC). A detailed search of the literature was made using PubMed, Cochrane Library, Excerpta Medica Database (Embase), China National Knowledge Infrastructure (CNKI) and Web of Science for related research publications written in English and Chinese. The methodological quality of the studies was also evaluated. The data were extracted and assessed by two reviewers independently. Analyses of the pooled data were performed. Odds Ratios (ORs) were calculated summarized. A final analysis of 1,125 CRC patients from 13 eligible studies was performed. The pooled OR from 13 studies was OR=37.31 (Confidence Interval (CI)=22.06-63.12, p<0.00001). Aberrant TFPI2 hyper-methylation was significantly higher CRC than in normal control. This present study observed that TFPI2 hyper-methylation was significantly higher in CRC than in normal controls, which suggest that TFPI2 hyper-methylation is associated with an increased risk of CRC.
Keywords
TFPI2, Methylation, Odds ratio.
Introduction
Colorectal Cancer (CRC) is one of the most malignant types of cancers. In the developed countries, CRC is the second most frequent cancer that leads to death [1]. Currently, there are about 1.25 million patients diagnosed with CRC, and more than 600,000 patients would die from CRC every year in the world [2]. Surgery can be used to extract the tumor if no lymph-node or distant metastasis, while the recurrence rate after surgery remains high [3]. Therefore, investigation of the mechanism of initiation and progression and identification of diagnostic markers are still needed in the treatment of CRC.
Tissue factor pathway inhibitor 2 is a protein that in humans is encoded by the TFPI2 gene. It is a serine protease inhibitor and plays an important role in the degradation and reshaping of Extracellular Matrix (ECM) [4]. The degradation and reshaping of ECM is the basic steps of tumor growth and metastasis, angiogenesis, cell migration, and normal physiological processes [5]. Studies have shown that with the progress of the breast cancer, colon cancer, pancreatic cancer, stomach cancer, laryngeal cancer, kidney cancer and other malignant tumor, the expression of TFPI2 is declining, and thus enhances the aggressivity of tumors. Other studies have confirmed that down-regulated expression of TFPI2 may have closely related with its promoter methylation. Therefore, DNA methylation of TFPI2 may promote tumor cell migration [6]. However, its role in CRC and its clinical significance have not been thoroughly investigated. The present study reviews and updates the published clinical investigations regarding the effect of TFPI2 promoter methylation on patients with CRC.
Methods
Search strategy and selection criteria
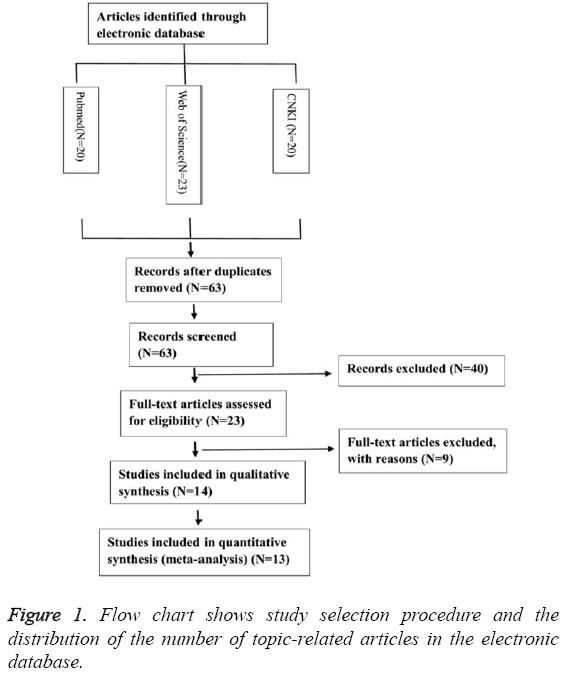
Searching PubMed, Cochrane Library, Excerpta Medica Database (Embase), China National Knowledge Infrastructure (CNKI) and Web of Science to identify studies by twelve 2015 using the search terms (“colorectal cancer” or “colon cancer” or “colorectal carcinoma” “colorectal tumor” or “colon tumor”) and (“hypermethylation” or “epigenetic” or “methylation”) and (“tissue factor pathway inhibitor 2” or “TFPI2” or “TFPI-2” or “REF1” or “PP5”). Conference abstracts were not selected for this analysis if they contain insufficient data. After the exclusion of redundant and/or irrelevant publications from the different databases, the fulltext versions of the remaining papers were estimated against exclusion and inclusion criteria (see the following paragraph), their reference lists searched for additional relevant articles. All searched data were retrieved twice.
The four inclusion criteria were as follows: 1) original study and the diagnosis of CRC was based on histopathology, 2) TFPI2 methylation evaluated in the circulation and/or primary CRC, 3) research revealed the relationship between TFPI2 methylation and CRC clinicopathologic parameters, 4) the subjects in every study comprised of CRC patients and healthy controls. The fore exclusion criteria were: 1) letters, reviews, case reports, conference abstracts, editorials, expert opinion; 2) not human studies; 3) not CRC studies; 4) and all in vitro/ex vivo studies.
Data extraction and methodological assessment
Two authors independently reviewed and extracted the data from eligible studies. Decisions were made, and disagreements about study selection were resolved, by discussion with a third reviewer. The two authors reviewed all of articles that fitted the inclusion criteria. The following information was recorded for each study: the first author’s name, year of publication, sample source, number of cases, clinicopathologic parameters, methylation detection method, methylation rate, and follow-up. Data for study characteristics and clinical responses were summarized and transferred into table format. The heterogeneity of investigation was evaluated to determine whether the data from the various studies could be analysed in a meta-analysis.
Statistical analysis
Analysis was conducted using R Software (R version 3.1.2). The strength of the association between TFPI2 gene promoter methylation and CRC was measured using a pooled Odds Ratio (OR) with a 95% Confidence Interval (CI). The frequency of TFPI2 hyper-methylation in different clinicopathologic features was compared. Heterogeneity among studies was evaluated with Cochran’s Q test [7] and the I2 statistic [8,9]. When heterogeneity was not an issue (I2<50%), a fixed-effects model was used to calculate parameters. If there was substantial heterogeneity (I2>50%), a random-effects model was used to pool data and attempt to identify potential sources of heterogeneity based on subgroup analyses. The pooled OR was estimated for the association between TFPI2 hyper-methylation and clinicopathologic features. P-values less than 0.05 were considered statistically significant.
Publication bias was assessed using a method reported by Begger’s funnel plot and Egger’s test [10]. Subgroup analysis and sensitivity analysis was used to explored the reasons of statistical heterogeneity.
Results
Identification of relevant studies
Thirty-three publications were identified using the described search method. Of these, 39 were excluded due to being laboratory studies, non-original articles (i.e., reviews), or irrelevant to the current analysis. Eventually, 13 studies were included in the final meta-analysis [11-23], as shown in Figure 1.
Study characteristics
Thirteen studies eligible for meta-analysis were published between 2009 and 2015, and between them reported on a total of 1,125 CRC patients from China, Japan, Korea, Germany, Netherlands, and the USA. Fifty-two point six two per cent of CRC patients had the methylated TFPI2 allele with a frequency ranging from 18.13% to 100% in individual trials. However, there was 9.23% of normal had the methylated TFPI2 allele with a frequency ranging from 0.00% to 50.00% in individual trials. The basic characteristics of these patients are summarized in Table 1. Among the 13 articles, 3 articles [18,20,22] focus on the clinicopathologic features of TNM staging system articles, the gender status and histology of CRC at the same time.
| Author | Year | Country | Sample | Method | Mean age (year) | TNM.stage | Male/Female | M1 | T1 | % | M2 | T2 | % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gerecke et al. [11] | 2015 | Germany | Tissue | MSP | 71.7 | NA | NA | 64 | 104 | 61.54% | 0 | 9 | 0.00% |
| Kim et al. [12] | 2013 | Korea | Tissue | QMSP | 65.8 | II-IV | NA | 8 | 8 | 100.00% | 18 | 36 | 50.00% |
| Zou et al. [13] | 2012 | USA | Tissue | QMSP | NA | NA | NA | 56 | 62 | 90.32% | 0 | 29 | 0.00% |
| Zhang et al. [14] | 2012 | China | Tissue | MSP | 60.8 | I-II | 50/30 | 52 | 80 | 65.00% | 0 | 30 | 0.00% |
| Hibi et al. [15] | 2012 | Japan | Serum | QMSP | NA | II-IV | NA | 39 | 215 | 18.14% | 0 | 20 | 0.00% |
| Ahlquist et al. [16] | 2012 | USA | Stool | QMSP | 69 | I-IV | 28/24 | 45 | 52 | 86.54% | 13 | 49 | 26.53% |
| Ahlquist et al. [16] | 2012 | USA | Serum | QMSP | 69 | I-IV | 28/24 | 31 | 52 | 59.62% | 3 | 46 | 6.52% |
| Zhang et al. [17] | 2011 | China | Stool | MSP | 52.5 | I-IV | 37/23 | 45 | 60 | 75.00% | 4 | 30 | 13.33% |
| Xu et al. [18] | 2011 | China | Stool | MSP | NA | I-IV | 37/23 | 42 | 60 | 70.00% | 1 | 30 | 3.33% |
| Kim et al. [19] | 2011 | Korea | Tissue | Chip | 63 | I-IV | 16-Jun | 7 | 22 | 31.82% | 0 | 22 | 0.00% |
| Hibi et al. [20] | 2011 | Japan | Serum | QMSP | 58 | I-IV | 125/90 | 39 | 215 | 18.14% | 0 | 20 | 0.00% |
| Xie et al. [21] | 2010 | China | Stool | MSP | 57 | I-IV | 17/13 | 19 | 30 | 63.33% | 0 | 30 | 0.00% |
| Hibi et al. [22] | 2010 | Japan | Tissue | QMSP | 66 | I-IV | 26/24 | 31 | 50 | 62.00% | 0 | 50 | 0.00% |
| Glockner et al. [23] | 2009 | Netherlands | Tissue | MSP | 65.2 | I-IV | 60/40 | 114 | 115 | 99.13% | 0 | 22 | 0.00% |
Table 1: Characteristics of eligible studies considered in the report.
Meta-analysis
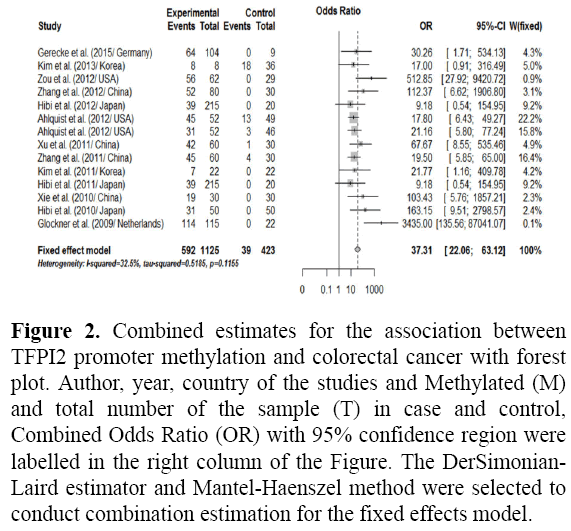
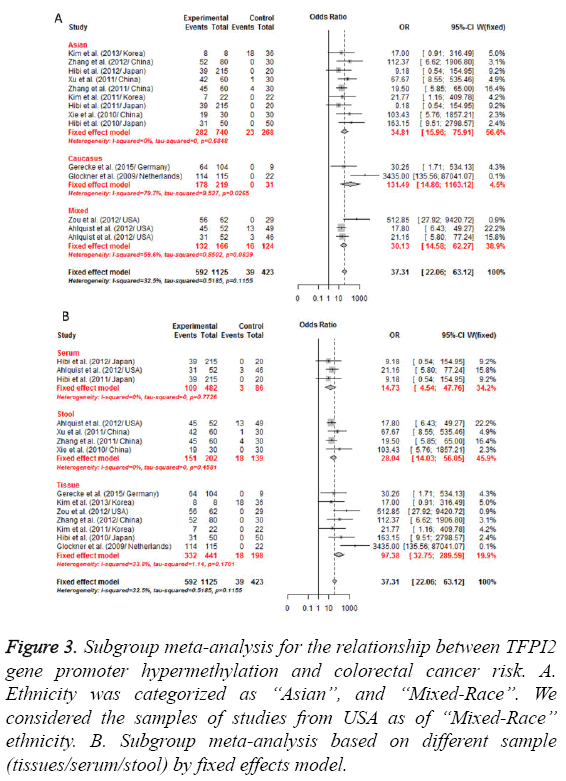
The results of the association between the promoter methylation of the TFPI2 and CRC and the heterogeneity test were shown in Figure 1. For TFPI2 promoter methylation associate with the CRC risk, this meta-analysis demonstrated that the frequency of TFPI2 promoter methylation in CRC was significantly higher than that normal control (cancer vs. normal: OR=37.31, 95% CI: 22.06 63.12, p<0.0001) with no heterogeneity (p=0.12) as shown in Figure 2. Subgroup analysis by ethnicity demonstrated that TFPI2 gene methylation was positively associated with the risk of CRC among between Asians, Mixed and Caucasians (all p<0.0001) when compared to healthy control as shown in Figure 3A. By comparing CRC to normal control as regards TFPI2 methylation in tissues/serum/stool the result found significant differences as shown in Figure 3B among tissues/serum/stool with association of TFPI2 methylation in CRC patients. In addition, subgroup analysis by methods used showed that the OR was 58.58 (95% CI=(25.88; 132.58), p<0.0001) in Methylation-Specific Polymerase Chain Reaction (MSP), 27.64 (95% CI=(14.01; 54.55), p<0.0001) in Quantitative Methylation Specific Polymerase Chain Reaction (QMSP).
Figure 2: Combined estimates for the association between TFPI2 promoter methylation and colorectal cancer with forest plot. Author, year, country of the studies and Methylated (M) and total number of the sample (T) in case and control, Combined Odds Ratio (OR) with 95% confidence region were labelled in the right column of the Figure. The DerSimonian- Laird estimator and Mantel-Haenszel method were selected to conduct combination estimation for the fixed effects model.
Figure 3: Subgroup meta-analysis for the relationship between TFPI2 gene promoter hypermethylation and colorectal cancer risk. A. Ethnicity was categorized as ‘‘Asian”, and ‘‘Mixed-Race’’. We considered the samples of studies from USA as of ‘‘Mixed-Race’’ ethnicity. B. Subgroup meta-analysis based on different sample (tissues/serum/stool) by fixed effects model.
By comparing early stage to advanced stage patients (cut of point was stage I and II vs. stage III and IV) as regards TFPI2 methylation in CRC of the meta result which were found a significant differences between both groups with association TFPI2 methylation in CRC with advanced stage patients. Meanwhile, the result of the histology was significantly, which was similarly to the result of TNM staging system. When comparison between patients with males and old females also showed a non-significant difference as regards methylation status in TFPI2 promoter as shown in Table 2. The heterogeneity was so big between the results of TNM staging system and histology as shown in Table 2.
| N | OR | 95% CI | p | Test of heterogeneity (p) | |
|---|---|---|---|---|---|
| Gender | 1.376 | (0.804; 2.356) | 0.245 | 0.184 | |
| Male | 188 | ||||
| Female | 137 | ||||
| Histology | 0.695 | (0.384; 1.257) | 0 | 0.229 | |
| Well | 258 | ||||
| Por | 67 | ||||
| TNM.stage | 0.421 | (0.243; 0.730) | 0.002 | 0.002 | |
| I-II | 111 | ||||
| III-IV | 174 |
Table 2: The relationship between TFPI2 methylation and difference clinical characteristics.
Sensitivity analyses and publication bias
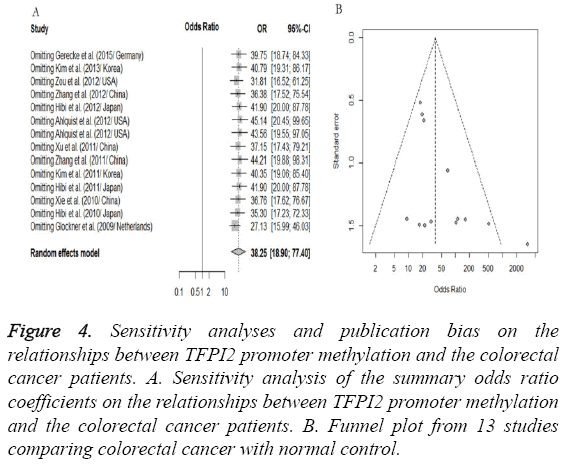
A “sensitivity analysis”, in which one by one study was removed at a time, was conducted to assess the stability of the results. The pooled ORs were not significantly changed when we removed one study, indicating the stability of our analyses as shown in Figure 4A. Similarly, the funnel plots were largely symmetrical as shown in Figure 4B which suggested there were no publication biases in this meta-analysis of TFPI2 methylation and CRC risk.
Figure 4: Sensitivity analyses and publication bias on the relationships between TFPI2 promoter methylation and the colorectal cancer patients. A. Sensitivity analysis of the summary odds ratio coefficients on the relationships between TFPI2 promoter methylation and the colorectal cancer patients. B. Funnel plot from 13 studies comparing colorectal cancer with normal control.
Discussion
CRC is a major cause of cancer-associated morbidity and mortality in the world. It is the second most prevalent cancer affects in males and females almost equally [24]. The epidemiologies of CRC have been studied by researchers but the result was insufficiency. Epigenetic alterations, particularly aberrant the methylations of DNA, one of the most primary epigenetic modifications, contribute to tumor initiation and progression. TFPI2 gene promoter region methylation has been reported involvement in the pathogenesis of Ovarian Cancer [24], prostate carcinoma [25] and colorectal cancer [20]. Here, a systematic review was made to access the association between the promoter methylation of the TFPI2 and CRC.
To our knowledge, this first meta-analysis of the relationship between of TFPI2 gene promoter methylation and CRC. This meta-analysis retrospectively studied 13 cohort studies aiming to investigate whether promoter DNA methylation of the TFPI2 gene has an impact on the risk of CRC. The results revealed that the frequency of TFPI2 gene promoter hypermethylation in CRC tissues/serum/stool significantly higher than that in normal tissues/serum/stool. Although the exact role of methylation status of the TFPI2 gene in CRC carcinogenesis is still indistinct, theory has been hypothesized that CpG islands hyper-methylation in the TFPI2 promoter region may lead to transcriptional silencing, and thus inhibit its function of tumor inhibition, resulting in the development of CRC. Subgroup analyses also were conducted to identify the relationships between TFPI2 gene promoter hyper-methylation and CRC risk. The results of subgroup analysis based on ethnicity demonstrated that TFPI2 gene promoter hypermethylation was closely associated with the risk of CRC among Asians, Caucasians and Mixed-race, revealing that there was no ethnic difference in the effects of TFPI2 gene promoter hyper-methylation on CRC risk. Considering the other limitations, such as the number of some published studies was not sufficiently large, the number of cases and controls was relatively small in the subgroup analyses, other pathological factors (such as the age status, smoking status, drinking status and environmental factors) can affect the frequency of DNA methylation. Hence, In order to provide a more precise estimation, well-designed studies are warranted by taking potential confounders such as smoking status, age status, drinking status and environmental factors into account.
For the difference clinicopathologic features, such as TNM staging system, histology and gender status, this meta-analysis result of 3 articles found that a significant difference in TNM staging system and histology, non-significant in gender status as regards methylation status in TFPI2 promoter. The same result was found by Hibi et al. [20,22]. However, the big heterogeneity was found in the result of TNM staging system, which were came from the small data or the difference clinical judgment standard that we suppose.
It is known that methylation is a major epigenetic modification in mammals, the stillness of the oncogene and tumor suppressor genes normally present in a dynamic equilibrium process, oncogene mutation or tumor-suppressor gene inactivation can directly or indirectly to break the balance, promote tumor formation and development. Hence, DNA methylation changes in cancer are well established, and translation of this knowledge toward screening has the potential biomarkers for significant health improvement. With the discovery of new disease screening markers, it may be possible to achieve early diagnosis, to have more informed choices for cancer treatment, and to identify markers associated with diagnosis and response to therapy.
Conclusion
Results of the present study show that DNA methylation of TFPI2 can down-regulated expression of TFPI2 and thus enhance the aggressivity of tumors. Similarly, this metaanalysis shows that TFPI2 hyper-methylation is significantly higher in CRC than in normal control, which suggests that the expression of TFPI2 is declining and TFPI2 hyper-methylation is associated with an increased risk of CRC.
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Bernard WSCPW. World cancer report 2014. International Agency for Research on Cancer 2014.
- Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev 2010; 19: 1893-1907.
- Weitz J, Koch M, Debus J, Hohler T, Galle PR. Colorectal cancer. Lancet 2005; 365: 153-165.
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T. The genomic landscapes of human breast and colorectal cancers. Science 2007; 318: 1108-1113.
- Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Meth 2008; 72: 124-132. Steiner FA, Hong JA, Fischette MR, Beer DG, Guo ZS, Chen GA. Sequential 5-Aza 2-deoxycytidine/depsipeptide FK228 treatment induces tissue factor pathway inhibitor 2 (TFPI-2) expressions in cancer cells. Oncogene 2005; 24: 2386-2397.
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177-188.
- Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557-560.
- DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med 1996; 15: 1237-1248.
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997; 315: 629-634.
- Gerecke C, Scholtka B, Lowenstein Y, Fait I, Gottschalk U, Rogoll D.Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is increased in inflamed colon tissue: putative risk markers for colitis-associated cancer. J Cancer Res ClinOncol 2015.
- Kim TO, Park J, Kang MJ, Lee SH, Jee SR. DNA hypermethylation of a selective gene panel as a risk marker for colon cancer in patients with ulcerative colitis. Int J Mol Med 2013; 31: 1255-1261.
- Zou H, Allawi H, Cao X, Domanico M, Harrington J. Quantification of methylated markers with a multiplex methylation-specific technology. ClinChem 2012; 58: 375-383.
- Zhang J, Yang S, Xie Y, Chen X, Zhao Y, He D. Detection of methylated tissue factor pathway inhibitor 2 and human long DNA in fecal samples of patients with colorectal cancer in China. Cancer Epidemiol 2012; 36:73-77.
- Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H. Methylation of TFPI2 no longer detected in the serum DNA of colorectal cancer patients after curative surgery. Anticancer Res 2012; 32:787-790.
- Ahlquist DA, Taylor WR, Mahoney DW, Zou H, Domanico M. The stool DNA test is more accurate than the plasma septin 9 test in detecting colorectal neoplasia. ClinGastroenterolHepatol 2012; 10: 272-277.
- Zhang JP, Wang J, Gui YL, Zhu QQ, Xu ZW.Human stool vimentin, oncostatin M receptor and tissue factor pathway inhibitor 2 gene methylation analysis for the detection of colorectal neoplasms. Zhonghua Yi XueZaZhi 2011; 91: 2482-2484.
- Xu ZW, Li JS, Zhang JP. Detection of OSMR and TFPI2 gene methylation in stool DNA for diagnosis of colorectal cancer. W Chinese J Digestol 2011; 19:1950-1953.
- Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, Min BH. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann SurgOncol 2011; 18:2338-2347.
- Hibi K, Goto T, Shirahata A, Saito M, Kigawa G, Nemoto H. Detection of TFPI2 methylation in the serum of colorectal cancer patients. Cancer Lett 2011; 311: 96-100.
- Xie YY, Li JS, Zhang JP, Yang SB. Human stool TFPI 2 gene methylation analysis in the diagnosis of colorectal cancer. Chinese J PractInt Med 2010; 7:642-643.
- Hibi K, Goto T, Kitamura YH, Yokomizo K, Sakuraba K, Shirahata A. Methylation of TFPI2 gene is frequently detected in advanced well-differentiated colorectal cancer. Anticancer Res 2010; 30:1205-1257.
- Glockner SC, Dhir M, Yi JM, McGarvey KE, Van Neste L. Methylation of TFPI2 in stool DNA: a potential novel biomarker for the detection of colorectal cancer. Cancer Res 2009; 69: 4691-4699.
- Bernard W. Stewart CPW. World Cancer Report . WHO 2014.
- Wang B, Yu L, Yang GZ, Luo X, Huang L. Application of multiplex nested methylated specific PCR in early diagnosis of epithelial ovarian cancer. Asian Pac J Cancer Prev 2015; 16:3003-3007.
- Ribarska T, Ingenwerth M, Goering W, Engers R, Schulz WA. Epigenetic inactivation of the placentally imprinted tumor suppressor gene TFPI2 in prostate carcinoma. CancGenomProteom 2010; 7:51-60.