Research Article - Biology & Medicine Case Reports (2018) Volume 2, Issue 2
A computational approach to identify microRNA (miRNA) based biomarker from the regulation of disease pathology
- *Corresponding Author:
- Anandaram H
Department of Bioinformatics, Sathyabama University, Chennai, India.
Tel: 9940066227
E-mail: harishchander.a@gmail.com
Accepted date: April 09, 2018
Citation: Anandaram H. A computational approach to identify microRNA (miRNA) based biomarker from the regulation of disease pathology. Biol Med Case Rep. 2018;2(2):12-25.
DOI: 10.35841/biology-medicine.2.2.12-25
Visit for more related articles at Biology & Medicine Case ReportsAbstract
In the post genomic era, identification of a potential miRNA in a computational approach for the significance of a discovery of systemic biomarker to treat diseases is a challenging task to execute. The challenge was addressed by identifying the associate genes from Pubmed, OMIM and DisgeNet and it was followed by identifying the miRNAs and transcription factors of associate target genes from RegNetworks. In the next step, a miRNA based regulatory network was constructed on the basis of association between gene-miR-TFs. Finally, the network was analyzed on the basis of statistical studies and miRNA based compatibility to identify a potential miRNA to be utilized as a biomarker to treat diseases in future. In this article, the computational approach was used for the identification of a miRNA based systemic biomarker in Psoriasis and in future this approach can also be used for other diseases.
Keywords
Post genomic era, RegNetworks, miRNA, Biomarkers.
Introduction
Psoriasis is a disorder mediated by immune system by making certain faulty signals in the human body. It’s still a belief that psoriasis can be developed under the specified condition i.e., “when the immune system signals the body to accelerate the growth of skin cells. In case of psoriasis, the skin cells mature in 3-6 days. Instead of being in shed, the cells in skin get pile up to cause the visible lesions. It was also found that the genes that cause psoriasis can determine the reaction of a person’s immune system. These genes can either cause psoriasis or other conditions which are immune-mediated like Type-I diabetes or rheumatoid arthritis. Pathophysiology of psoriasis involves the understanding of the occurrence of prominent pathologies in the major components of skin i.e., the epidermis and the dermis. There are two well established hypotheses about the process that occurs in the development of the disease. The first hypothesis considers psoriasis as a disorder with excessive growth and reproduction of skin cells. Here, the problem is viewed as a fault of the epidermis and its keratinocytes. In second hypothesis, the disease is viewed as an immune-mediated disorder. Here, the excessive reproduction of skin cells is secondary to the factors produced by the immune system [1,2].
Micro RNA is a family of non-coding RNA (ncRNA) which was discovered in 1993, it consist of 19-25 nucleotides and regulates the expression of approximately 30% of proteincoding miRNAs in humans [3]. Base pairing at the position of 2-8 nucleotides were relative to the 5′ end of the small RNA to be termed as the “seed” region and it appears to be important for target recognition. Maturation of miRNAs involves multiple steps and initially two intermediate forms of miRNAs, namely primary (pri-) and precursor (pre-) miRNAs, were produced sequentially. In this process, Drosha (RNase III enzyme) and the double-stranded RNA (dsRNA) binding protein Dgcr8 cleaves the pri-miRNAs to produce a hairpin-shaped pre-miRNAs that are recognized by Exportin5 and they are subsequently transported from the nucleus to cytoplasm. There is another RNase III enzyme called Dicer which cleaves the pre-miRNAs to release ~22-nt double-stranded RNA duplexes (namely miRNA/miRNA* duplexes) with ~2-nt 3’ overhangs [4]. One strand of a RNA duplex is termed as a mature miRNA which is further loaded into an Argonaute protein in the RNA-induced silencing complex (RISC) to exert its regulatory function on the basis of its binding with the target transcripts [5].
A unique miRNA can regulate the expression of hundreds of proteins and the expression of a specific protein may be controlled by several miRNAs [6]. The sequence conservation of most miRNAs lies between the distantly related organisms to suggest the impact of a strong evolutionary pressure [7] and they have been shown to participate in many fundamental life processes like development, differentiation, organogenesis, growth control and apoptosis. Accordingly, deregulation of miRNA expression has been shown to contribute to cancer, heart diseases, infectious diseases, inflammatory diseases and other medical conditions, making them potential targets for medical diagnosis and therapy [8]. Initially, Lee had found lin-4 as a regulator of developmental timing in nematode Caenorhabditis elegans [9]. After several years, Reinhart had discovered lethal-7 (let-7) gene in Caenorhabditis elegans [10]. At present, 2500 miRNAs are in the human genome. Majority of miRNA are intragenic [11]. Micro RNAs are initially transcribed as a part of an RNA stem-loop that in turn forms part of a several hundred nucleotides long miRNA precursor miRNA (primiRNA) [12-15].
Materials and Methods
PubMed
PubMed is an online search engine with open access facility to refer MEDLINE for identifying references and abstracts on topics in biomedical and life sciences. The United States National Library of Medicine (NLM) at the National Institutes of Health maintains the database as part of the Entrez system to retrieve information. Most of the records in PubMed contain links to the complete article, in PubMed Central [16-18]. Information regarding the indexed journals in MEDLINE can be found in the Catalog of NLM.
DisGeNET
DisGeNET [19] is a platform of pattern discovery, designed for addressing the queries regarding the genetic imprint of human diseases. DisGeNET is one of the largest repositories of genedisease associations (GDAs) in humans [19]. It offers a set of tools in bioinformatics to facilitate the data analysis by different users. It is maintained by the Integrative Biomedical Informatics (IBI) Group of the (GRIB)-IMIM/UPF at the Barcelona Biomedical Research Park (PRBB), Barcelona in Catalonia.
OMIM
Online Mendelian Inheritance in Man (OMIM) is a comprehensive compendium of human genes and phenotypes [20] that are available freely and updated daily. The complete text, referenced in the overviews of OMIM contains information on all known Mendelian disorders for 15,000 genes.
RegNetwork
RegNetwork [21] is a data base that contains five types (Transcription factor-transcription factor, transcription factor-gene, transcription factor-microRNA, and microRNAtranscription factor) of transcriptional and posttranscriptional regulatory relationships for human and mouse.
Cytoscape
Cytoscape [22] software is used for network construction, visualization and analysis in bioinformatics with an open source platform for visualizing the interactions in molecular networks and integrating them with the profiles of gene expression. Additional features in cytoscape are available as plugins for network and molecular profiling.
Cytohubba
Cytohubba [23] is a cytoscape plugin for performing the analyses of gene regulation and protein-protein interaction involved in the process of cellular pathways in the process of signal transduction. Cytohubba ranks the nodes of network by topological methods like radiality, betweenness, closeness, bottleneck, eccentricity and etc.
MiRmap
miRmap [24] software addresses the challenges in post transcriptional repression of miRNAs in human genome by evolutionary, probabilistic thermodynamic and sequencebased features.
Triplex RNA
Triplex RNA [25] is a database of cooperating micrRNAs with their mutual targets. In this database miRNA target prediction is based on the analysis of predicted miRNA triplex with molecular dynamics simulations and differential modeling procedures in mathematics.
DAVID
The Database for Annotation, Visualization and Integrated Discovery (DAVID) contain complete information about functional annotation of genes. The current version of DAVID [26] is 6.8 and it provides a set of comprehensive tools for functional annotation of genes.
Methodology (Computational approach of miRNA associated regulation)
1. Identify the disease associated genes from Pubmed, DisGeNET and OMIM.
2. Obtain the associated list of miRNAs and transcription factors for the disease associated genes from Reg networks.
3. Construct and analyze the network in Cytoscape.
4. Identify the miRNA based hub genes and transcription factors from cytohubba.
5. Identify the implication of miRNA in Regulatory network in miRmap and miRNA triplex.
6. Identify and analyze the gene associated pathways in DAVID.
Results
Text mining of Genes from Pubmed, DisGeNET and OMIM along with the miRNAs and transcription factors from Regnetworks resulted in the identification of interaction between 92 genes-437 miRNA-285 transcription factors and the results were given in Table 1. A regulatory network was constructed in Cytoscape and the properties of the network were analyzed in the Network Analyzer. Finally, the hub genes were identified from cytohubba and the miRNA based regulation was analyzed on the basis of seed pairing in miRmap and the experimental evidences from the previous literature.
| S.No. | Genes (Pubmed, DisGeNET and OMIM) |
miRNAs (RegNetworks) |
Transcription Factor (RegNetworks) |
|---|---|---|---|
| 1. | HPSE | hsa-miR-1258 | ETS1; ETS2; ETV4; MAX; MXI1::CLEC5A; AR; ESR1 |
| HOXA7 | |||
| 2. | CCL20 | hsa-miR-21-5p; hsa-miR-144; hsa-miR-145; hsa-miR-21; hsa-miR-330-3p; hsa-miR-338-5p; hsa-miR-361-3p | CTCF;MYC; PPARG::RXRA |
| RELA; SP1 | |||
| hsa-miR-380; hsa-miR-496; hsa-miR-518e; hsa-miR-525-5p; hsa-miR-548d-3p; hsa-miR-590-3p; hsa-miR-590-5p | |||
| hsa-miR-635; hsa-miR-766; hsa-miR-802; hsa-miR-921 | |||
| 3. | CCL2 | hsa-miR-124-3p; hsa-miR-124; hsa-miR-124a; hsa-miR-141; hsa-miR-142-5p; hsa-miR-323-3p; hsa-miR-374a; hsa-miR-374b; hsa-miR-421; hsa-miR-495; hsa-miR-545; hsa-miR-577; hsa-miR-633 | JUN; NFIC; NFKB1; NFKB2; NR2F2; REL; RELA; SMAD3 SMAD4; SP1; SRF; STAT1; STAT2; STAT3; STAT4; STAT6 |
| 4. | EIF4E | hsa-miR-1; hsa-miR-122; hsa-miR-141; hsa-miR-145-3p; hsa-miR-146b-5p; hsa-miR-150; hsa-miR-16; hsa-miR-186; hsa-miR-195; hsa-miR-203; hsa-miR-206; hsa-miR-325; hsa-miR-34c-3p; hsa-miR-377; hsa-miR-380; hsa-miR-495; hsa-miR-498; hsa-miR-503; hsa-miR-520d-5p; hsa-miR-524-5p; hsa-miR-545; hsa-miR-582-5p; | BACH1; CUX1; EMX2; FOS; FOSB; FOSL1; FOXD1; FOXF2; JUN; JUNB; JUND; MAX; MXI1::CLEC5A; MYC |
| NFIL3; NFYA; NR3C1; PML | |||
| STAT1; STAT2; STAT3; STAT4; STAT6; USF1 | |||
| hsa-miR-586; hsa-miR-592; hsa-miR-599; hsa-miR-613; hsa-miR-654-5p; hsa-miR-656; has-miR-9; has-miR-141-3p; has-miR-145-5p; has-miR-497-5p; has-miR-768-3p | |||
| 5. | PPARD | hsa-miR-138-5p; hsa-miR-29b; hsa-miR-29c; hsa-miR-93 | ATF1; ATF2; ATF3; ATF4; ATF5; ATF6; ATF7; BCL6; CREB1; CTCF; EGR1; EP300; GABPA; HDAC1; HDAC2; HDAC3; HDAC7; JUP; LEF1; NCOR1; NCOR2; NR0B2; NRIP1; PROX1; RELA; RXRA; RXRB; RXRG; SMAD9; SPEN |
| TCF7; TCF7L1; TCF7L2 | |||
| 6. | TAP2 | hsa-miR-330-3p; hsa-miR-370-3p; hsa-miR-384; hsa-miR-670-3p; hsa-miR-6893-3p; hsa-miR-185; hsa-miR-219-2-3p; hsa-miR-330-3p; hsa-miR-370; hsa-miR-371-5p; hsa-miR-384; | CREB1; CUX1; ESR1; MAX; MYC; NFE2L1; STAT5A |
| hsa-miR-409-3p; hsa-miR-433; hsa-miR-522; hsa-miR-582-5p; hsa-miR-645; hsa-miR-655; hsa-miR-875-3p; hsa-miR-885-5p; hsa-miR-921 | |||
| 7. | CYLD | hsa-miR-181b-5p; hsa-miR-182-5p; hsa-miR-362-5p; hsa-miR-500a-5p | ATF2; EGR1; IKBKG; JUN; LHX3; NFKB1; NFYA; POU2F1; SP1; TCF3 |
| hsa-miR-130a; hsa-miR-130b; hsa-miR-15a; hsa-miR-15b; hsa-miR-16 hsa-miR-181b; hsa-miR-181d; hsa-miR-182; hsa-miR-186; hsa-miR-195 |
|||
| hsa-miR-197; hsa-miR-19a; hsa-miR-19b; hsa- | |||
| miR-301; hsa-miR-301a; hsa-miR-301b; hsa-miR-340; hsa-miR-362-5p; hsa-miR-424; hsa-miR-454 | |||
| hsa-miR-497; hsa-miR-508-3p; hsa-miR-543; hsa-miR-544; hsa-miR-548a-5p; hsa-miR-548b-5p; hsa-miR-548c-5p; hsa-miR-548d-5p; hsa-miR-579; hsa-miR-590-3p; hsa-miR-656; | |||
| hsa-miR-944 | |||
| 8. | IGF1 | hsa-miR-27a-3p; hsa-miR-29a-3p; hsa-miR-190a-5p; hsa-miR-199a-3p; hsa-let-7i-5p; hsa-miR-299-3p; hsa-miR-190b; hsa-let-7e-5p; hsa-miR-483-3p; hsa-let-7a; hsa-let-7b; hsa-let-7c; hsa-let-7e; hsa-let-7f; hsa-let-7g; hsa-let-7i; hsa-miR-1; hsa-miR-105; hsa-miR-128; hsa-miR-129-5p; hsa-miR-1297; hsa-miR-130a; hsa-miR-130b; hsa-miR-149; hsa-miR-152; hsa-miR-154; hsa-miR-186; hsa-miR-18a; hsa-miR-18b; hsa-miR-190; hsa-miR-192; hsa-miR-19a; hsa-miR-19b; hsa-miR-206; hsa-miR-215; hsa-miR-221; hsa-miR-222; hsa-miR-23a; hsa-miR-23b; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-301a; hsa-miR-301b; hsa-miR-340; hsa-miR-361-5p; hsa-miR-362-5p; hsa-miR-377; hsa-miR-410; hsa-miR-425; hsa-miR-450b-5p; hsa-miR-454; hsa-miR-486-5p; hsa-miR-488; hsa-miR-489; hsa-miR-495; hsa-miR-502-5p; hsa-miR-503; hsa-miR-519a; hsa-miR-519b-3p; hsa-miR-519c-3p; hsa-miR-556-3p; hsa-miR-561; hsa-miR-576-5p; hsa-miR-590-3p; hsa-miR-592; hsa-miR-599; hsa-miR-607; hsa-miR-613; hsa-miR-626; hsa-miR-632; hsa-miR-634; hsa-miR-656; hsa-miR-758;hsa-miR-942; hsa-miR-98 | CEBPA; CTCF; ESR1; FOXD1; FOXF2; JUN; MYB; POU3F2; RFX1; STAT1; STAT2; STAT3; STAT5A; STAT5B; TGIF1 |
| 9. | BCL2 | hsa-miR-34b-5p; hsa-miR-21-5p; hsa-miR-153-3p; hsa-miR-204-5p; hsa-let-7a-5p; hsa-miR-15a-5p; hsa-miR-15b-5p; hsa-miR-16-5p; hsa-miR-34a-5p; hsa-miR-20a-5p; hsa-miR-17-5p; hsa-miR-29a-3p; hsa-miR-29b-3p; hsa-miR-29c-3p; hsa-miR-34b-3p; hsa-miR-181a-5p; hsa-miR-181b-5p; hsa-miR-181c-5p; hsa-miR-181d-5p; hsa-miR-33b-5p; hsa-miR-34c-5p; hsa-miR-192-5p; hsa-miR-195-5p; hsa-miR-630; hsa-miR-451a; hsa-miR-125b-5p; hsa-miR-365a-3p; hsa-miR-449a; hsa-miR-200b-3p; hsa-miR-200c-3p; hsa-miR-429; hsa-miR-7-5p; hsa-miR-136-5p; hsa-miR-24-2-5p; hsa-miR-148a-3p; hsa-miR-24-2-5p; hsa-miR-182-5p; hsa-miR-143-3p; hsa-miR-205-5p; hsa-miR-126-3p; hsa-miR-18a-5p; hsa-miR-497-5p; hsa-miR-1915-3p; hsa-miR-206; hsa-miR-448; hsa-miR-708-5p; hsa-miR-184; hsa-miR-30b-5p; hsa-miR-135a-5p; hsa-miR-224-5p; hsa-miR-503-5p; hsa-miR-494-3p; hsa-miR-211-5p | |
| 10. | AREG | hsa-miR-34a-5p; hsa-miR-200a-3p; hsa-miR-129-5p; hsa-miR-135a;hsa-miR-135b; hsa-miR-345; hsa-miR-34a; hsa-miR-34c-5p; hsa-miR-449a; hsa-miR-449b; hsa-miR-499-5p; hsa-miR-517a; hsa-miR-517c; hsa-miR-548a-5p; hsa-miR-548c-5p; hsa-miR-556-5p; hsa-miR-559; hsa-miR-561; hsa-miR-577; hsa-miR-583; hsa-miR-584; hsa-miR-590-3p; hsa-miR-640 | AR; BRCA1; CREB1; E2F1; EGR1; HOXB13; IRF1; NFKB1 PAX2; RARA; RELA; SMAD3 SMAD4; SP1; STAT5A; WT1 |
| hsa-miR-944 | |||
| 11. | VNN3 | hsa-miR-138-5p; hsa-miR-455-5p; hsa-miR-135a; hsa-miR-135b; hsa-miR-199a-3p; hsa-miR-345; hsa-miR-371-5p; hsa-miR-421; hsa-miR-455-3p; hsa-miR-455-5p; hsa-miR-505 | CTCF; FOXA2; NFKB1;NFKB2 REL; RELA; STAT5B |
| hsa-miR-514; hsa-miR-744 | |||
| 12. | MMP9 | hsa-miR-451a; hsa-miR-491-5p; hsa-miR-338-3p; hsa-miR-9-5p; hsa-miR-211-5p; hsa-let-7e-5p; hsa-miR-133b; hsa-miR-29b-3p; hsa-miR-191; hsa-miR-204; hsa-miR-339-5p; hsa-miR-451; hsa-miR-483-3p; hsa-miR-494 hsa-miR-515-5p; hsa-miR-520a-5p; hsa-miR- |
AR; BACH1; BACH2; ERG; ETS1; ETS2; ETV4; FLI1; FOS; FOSB; FOSL1; JUN; JUNB; JUND; MYC; NFE2; NFE2L1; NFKB1; NFKB2; PPARA; PPARG; RELA; RELB; SMAD3; SP1; SPI1 |
| 525-5p | |||
| 13. | HBEGF | hsa-miR-194-5p; hsa-miR-132-3p; hsa-let-7d; hsa-let-7g; hsa-let-7i; hsa-miR-132; hsa-miR-135a; hsa-miR-135b; hsa-miR-182; hsa-miR-183; hsa-miR-194; hsa-miR-212; hsa-miR-27a; hsa-miR-27b; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-31; hsa-miR-376c; hsa-miR-379; hsa-miR-623; hsa-miR-662; hsa-miR-96 | CTCF; ETS2; MAX; TBP; ZBTB16 |
| 14. | TGFA | hsa-miR-152-3p; hsa-miR-376c-3p; hsa-miR-101; hsa-miR-130a; hsa-miR-130b; hsa-miR-137; hsa-miR-148a; hsa-miR-148b; hsa-miR-152; hsa-miR-205; hsa-miR-23a; hsa-miR-23b; hsa-miR-301 | CTCF; EPAS1; ESR1; FOXA1; FOXA2; HIF1A; NFKB1; NFKB2; NKD2; PGR; TFAP2A; TP53 |
| 15. | LHFP | hsa-miR-133a-3p.2; hsa-miR-133b; hsa-miR-101; hsa-miR-133a; hsa-miR-133b; hsa-miR-141; hsa-miR-147; hsa-miR-153; hsa-miR-200a; hsa-miR-200b; hsa-miR-200c; hsa-miR-218; hsa-miR-26a; hsa-miR-297; hsa-miR-300; hsa-miR-337-3p; hsa-miR-340; hsa-miR-381; hsa-miR-429; hsa-miR-448; hsa-miR-491-3p; hsa-miR-500; hsa-miR-501-5p; hsa-miR-607; hsa-miR-618; hsa-miR-632; hsa-miR-9 | AHR; ARNT; CTCF; PATZ1 |
| 16. | EGFR | hsa-miR-7-5p; hsa-miR-145-5p; hsa-miR-128-3p; hsa-miR-146a-5p; hsa-miR-21-5p; hsa-miR-128b; hsa-miR-133a-3p; hsa-miR-133b; hsa-miR-27a-3p; hsa-let-7a-5p; hsa-miR-574-3p; hsa-miR-219a-5p; hsa-miR-302b-3p; hsa-miR-125a-5p; hsa-miR-218-5p; hsa-miR-21; hsa-miR-1; hsa-miR-128; hsa-miR-141; hsa-miR-146a; hsa-miR-16; hsa-miR-21; hsa-miR-27a; hsa-miR-27b; hsa-miR-548c-3p; hsa-miR-7 | AR; CEBPB; CTNNB1; E2F1; EGR1; ELF3; ESR1; ESR1; ESR2; HOXC10; HTT; MEF2A; MYB; NFKB2; PPARG; REL; RELA; SMURF2; SP1; STAT1; STAT3; STAT5A; STAT5B; TFAP2A; TP53; WT1; WWP1; XRCC6 |
| 17. | SGCG | hsa-miR-137 | Nil |
| 18. | SDC4 | hsa-miR-18a-5p; hsa-miR-1; hsa-miR-124; hsa-miR-194; hsa-miR-224; hsa-miR-506; hsa-miR-548d-3p; hsa-miR-637; hsa-miR-941 | EBF1; MAX; NFKB1; REL; RELA; SP1; STAT5A; STAT5B; mTFAP2A; TFAP2C; TGFB1I1 |
| 19. | IGF1R | hsa-miR-122-5p; hsa-miR-133b; hsa-miR-145-5p; hsa-miR-7-5p; hsa-miR-138-5p; hsa-miR-194-5p; hsa-miR-99a-5p; hsa-miR-223-3p; hsa-miR-100-5p; hsa-miR-497-5p; hsa-miR-152-3p; hsa-miR-139-5p; hsa-miR-376a-3p; hsa-miR-376c-3p; hsa-miR-383-5p; hsa-miR-181b-5p; hsa-miR-335-5p; hsa-miR-320a; hsa-let-7e-5p; hsa-miR-125b-2-3p; hsa-let-7c-5p; hsa-miR-16-5p; hsa-miR-630; hsa-let-7b-5p; hsa-miR-143-3p; hsa-miR-133a-3p; hsa-miR-140-5p; hsa-miR-150-3p; hsa-miR-375; hsa-miR-503-5p; hsa-miR-378a-3p; hsa-miR-185-5p; hsa-miR-206; hsa-miR-21-5p; hsa-miR-26b-5p; hsa-miR-486-5p; hsa-let-7a; hsa-let-7b; hsa-let-7c; hsa-let-7d; hsa-let-7e; hsa-let-7f; hsa-let-7g; hsa-let-7i; hsa-miR-100; hsa-miR-106a; hsa-miR-122; hsa-miR-133b; hsa-miR-138; hsa-miR-139-5p; hsa-miR-140-5p; hsa-miR-141; hsa-miR-143; hsa-miR-145; hsa-miR-153; hsa-miR-15a; hsa-miR-15b; hsa-miR-16; hsa-miR-182; hsa-miR-186; hsa-miR-194; hsa-miR-195; hsa-miR-200a; hsa-miR-202; hsa-miR-203; hsa-miR-214; hsa-miR-22; hsa-miR-223; hsa-miR-300; hsa-miR-302b; hsa-miR-302c; hsa-miR-302d; hsa-miR-30a; hsa-miR-30b; hsa-miR-30c; hsa-miR-30d; hsa-miR-30e; hsa-miR-320; hsa-miR-328; hsa-miR-329; hsa-miR-330-3p; hsa-miR-340; hsa-miR-361-3p; hsa-miR-362-3p; hsa-miR-372; hsa-miR-373; hsa-miR-376c; hsa-miR-377; hsa-miR-378; hsa-miR-379; hsa-miR-381; hsa-miR-409-5p; hsa-miR-422a; hsa-miR-424; hsa-miR-448; hsa-miR-455-5p; hsa-miR-489; hsa-miR-493; hsa-miR-494; hsa-miR-495; hsa-miR-497; hsa-miR-503; hsa-miR-505; hsa-miR-507; hsa-miR-509-3-5p; hsa-miR-509-5p; hsa-miR-520a-3p; hsa-miR-520b; hsa-miR-520c-3p; hsa-miR-520d-3p; hsa-miR-520d-5p; hsa-miR-520e; hsa-miR-524-5p; hsa-miR-539; hsa-miR-548c-3p; hsa-miR-548d-3p; hsa-miR-557; hsa-miR-570; hsa-miR-577; hsa-miR-583; hsa-miR-625; hsa-miR-626; hsa-miR-646;hsa-miR-650; hsa-miR-653; hsa-miR-671-3p; hsa-miR-7; hsa-miR-769-5p; hsa-miR-892b; hsa-miR-944; hsa-miR-96; hsa-miR-98; hsa-miR-99a | BACH1; BACH2; BRCA1; CTNNB1; E2F1; E2F2; E2F3; E2F4; E2F5; E2F6; E2F7; EGR1; ESR1; FOXO1; FOXO3; FOXO3B; FOXO4; MAX; |
| MXI1::CLEC5A; MYB; MYC; MZF1; PAX5; REL | |||
| RELA; SMURF2; SP1; SREBF1; SREBF2; STAT3; TFAP2A; TP53 | |||
| USF1; WT1; WWP1 | |||
| 20. | LEP | hsa-miR-9-5p; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-331-3p | ARNT; CEBPA; CEBPB |
| CEBPD; FOXC1; HIF1A HLF;MIF;TBP |
|||
| hsa-miR-369-5p; hsa-miR-520g | |||
| hsa-miR-520h; hsa-miR-575; hsa-miR-875-5p; hsa-miR-9 | |||
| 21. | KLK13 | hsa-miR-330-3p; hsa-miR-455-5p; hsa-miR-542-3p; hsa-miR-591; hsa-miR-620; hsa-miR-654-5p | CREB1; EGR1; KLF12; MZF1; PPARG |
| 22. | HMOX1 | hsa-miR-196a-5p; hsa-miR-122-5p; hsa-miR-24-3p; hsa-miR-16; hsa-miR-196a-3p; hsa-miR-873 | BACH2; CREB1; ERG; ETS1; FLI1; HIF1A; HNF4A; MAX; MXI1::CLEC5A; MYC; NFE2; NFIC; NFKB1; PPARG::RXRA; RXRA; SMAD7; SP1; SPI1; STAT3; TFAP2A; USF1; USF2 |
| 23. | IFI6 | hsa-miR-1225-3p; hsa-miR-558; hsa-miR-624; hsa-miR-920 | TFAP2C; USF1 |
| 24. | SFXN1 | hsa-miR-30a-5p; hsa-miR-30b-5p; hsa-miR-30c-5p; hsa-miR-30d-5p; hsa-miR-30e-5p; hsa-miR-1; hsa-miR-128a | HNF4A; MAX; MYC |
| hsa-miR-134; hsa-miR-30a; hsa-miR-30b; hsa-miR-30c; hsa-miR-30d; hsa-miR-30e | |||
| 25. | IL23R | hsa-miR-383-5p.2; hsa-miR-216a; hsa-miR-297; hsa-miR-331-5p; hsa-miR-454; hsa-miR-509-3p; hsa-miR-583; hsa-miR-875-3p; hsa-miR-876-3p; hsa-miR-936 | FOS; JUN; RORA; STAT3 |
| 26. | PTPN22 | hsa-miR-181a-5p; hsa-miR-133a; hsa-miR-133b; hsa-miR-325; hsa-miR-630 | CDC5L; IRF1; MEF2A TP53 |
| 27. | LOR | hsa-miR-196a-5p; hsa-miR-196b-5p; | ATF1; CREB1; FOS; FOSB; JUN; JUNB; JUND; SP3 |
| hsa-let-7a; hsa-let-7b; hsa-let-7c; hsa-let-7d; hsa-let-7e; hsa-let-7f; hsa-let-7g | |||
| hsa-let-7i; hsa-miR-135a; hsa-miR-135b; hsa-miR-196a; hsa-miR-196b; hsa-miR-28-3p; hsa-miR-296-3p; hsa-miR-331-3p; hsa-miR-450b-5p; hsa-miR-490-5p; hsa-miR-570; hsa-miR-583; hsa-miR-641; hsa-miR-766; hsa-miR-873; hsa-miR-875-3p; hsa-miR-922; hsa-miR-98 | |||
| 28. | S100A9 | hsa-miR-196a-5p | AR; CTCF; MYB; RARA |
| RARB; RARG; SPI1; TBP | |||
| TFAP2A; TP53 | |||
| 29. | S100A8 | hsa-miR-24-3p; hsa-miR-135a; hsa-miR-135b; hsa-miR-202; hsa-miR-326 | AR; FOS; FOSB; JUN; JUNB; JUND; PDCD11; RARA; RARB; RARG |
| hsa-miR-330-5p; hsa-miR-544 | TBP; TP53 | ||
| 30. | IL10 | hsa-miR-106a-5p; hsa-let-7c-5p; hsa-let-7a; hsa-let-7b; hsa-let-7c; hsa-let-7d | ATF1; CEBPA; CEBPB; CREB1; E2F1; ESR1; ETS1; MEF2A; NFKB1 POU3F2; PPARG; SP1; STAT3; TBP |
| hsa-let-7e; hsa-let-7f; hsa-let-7g; hsa-let-7i; hsa-miR-106a; hsa-miR-10b; hsa-miR-142-3p; hsa-miR-186; hsa-miR-198; hsa-miR-202; hsa-miR-337-5p; hsa-miR-543; hsa-miR-588; hsa-miR-597; hsa-miR-630; hsa-miR-671-5p; hsa-miR-769-5p; hsa-miR-888; hsa-miR-98 | |||
| 31. | IL24 | hsa-miR-203a-3p; hsa-miR-205-5p; hsa-miR-132; hsa-miR-140-3p;hsa-miR-141; hsa-miR-183; hsa-miR-186; hsa-miR-200a; hsa-miR-200b; hsa-miR-200c; hsa-miR-203; hsa-miR-205 | CEBPB; JUN; TFAP2A |
| TFAP2C | |||
| hsa-miR-27a; hsa-miR-27b; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-300; hsa-miR-324-3p; hsa-miR-338-5p; hsa-miR-380; hsa-miR-381; hsa-miR-425; hsa-miR-429; hsa-miR-452; hsa-miR-495; hsa-miR-506; hsa-miR-518a-3p; hsa-miR-518b; hsa-miR-518c; hsa-miR-518f; hsa-miR-520a-5p; hsa-miR-525-5p; hsa-miR-573; hsa-miR-582-5p; hsa-miR-600; hsa-miR-601; hsa-miR-602; hsa-miR-616; hsa-miR-628-5p; hsa-miR-767-5p | |||
| hsa-miR-891b; hsa-miR-943 | |||
| 32. | ADAM17 | hsa-miR-26a-5p; hsa-miR-122-5p; hsa-miR-145-5p; hsa-miR-152-3p | CTCF; EGR1; FHL2; GABPA; HNF4A; NOTCH2; NOTCH3; NOTCH4;PPARG::RXRA; |
| SP1; TFAP2A; TFAP2C; YY1 | |||
| 33. | IL36RN | hsa-miR-216a-5p; hsa-miR-122; hsa-miR-338; hsa-miR-338-3p; hsa-miR-507; hsa-miR-197; hsa-miR-338-3p | MCM6; SKIL; SSBP2 |
| SSBP4 | |||
| 34. | IL1RN | hsa-miR-125a-5p; hsa-miR-125b-3p; hsa-miR-371-3p; hsa-miR-515-5p | BACH1; BACH2; EBF1; FOXA2; HNF4A; NFKB1; |
| NR3C1; PAX5; RXRA; RXRB::RARB; SPI1; STAT5A | |||
| 35. | CTLA4 | hsa-miR-155-5p; hsa-miR-101; hsa-miR-105; hsa-miR-155; hsa-miR-205 | BPTF; STAT5A; STAT5B |
| hsa-miR-380; hsa-miR-384; hsa-miR-429; hsa-miR-432; hsa-miR-449b; hsa-miR-451; hsa-miR-496; hsa-miR-516a-3p; hsa-miR-517a; hsa-miR-651 | |||
| hsa-miR-656 | |||
| 36. | SGPP2 | hsa-miR-101-3p.1; has-miR-24 | EBF1; NFKB1; NFKB2; RELA; RELB; TCF3; ZEB1 |
| 37. | IRF2 | hsa-miR-20a-5p; hsa-miR-153; hsa-miR-18a; hsa-miR-18b; hsa-miR-214 | EP300; HMGN1; IRF2BP1;IRF7; IRF8; KAT2B; MAX; MXI1::CLEC5A;MYC; NFKB1; NFKB2;RELA; RELB; STAT1 |
| hsa-miR-220c; hsa-miR-221; hsa-miR-222; hsa-miR-23a; hsa-miR-23b; hsa-miR-26a; hsa-miR-26b; hsa-miR-302a | |||
| hsa-miR-302b; hsa-miR-302c; hsa-miR-302d; hsa-miR-340; hsa-miR-342-5p; hsa-miR-372; hsa-miR-373; hsa-miR-455-5p; hsa-miR-495; hsa-miR-512-3p; hsa-miR-520a-3p; hsa-miR-520b; hsa-miR-520c-3p; hsa-miR-520d-3p; hsa-miR-520e; hsa-miR-520f; hsa-miR-549; hsa-miR-553 | |||
| hsa-miR-556-5p; hsa-miR-568; hsa-miR-571; hsa-miR-574-5p; hsa-miR-648; hsa-miR-934 | |||
| 38. | IL4 | hsa-miR-340-5p; hsa-miR-410-3p; | CEBPA; CEBPB; CEBPG; ETV4; GATA1; NFKB1; POU2F1; POU2F2; RELA; |
| hsa-miR-29a | |||
| STAT1; STAT2; TFAP2A | |||
| TP53 | |||
| 39. | IL12B | hsa-miR-23a-3p; hsa-miR-23b-3p; hsa-miR-23c; hsa-miR-130a-5p; hsa-miR-183; hsa-miR-219-5p; hsa-miR-220c | CEBPA; CEBPB; ETS1; ETS2; FOS; IRF5; JUN; NFKB1; REL; RELA; SP1 |
| hsa-miR-494; hsa-miR-545; hsa-miR-632; hsa-miR-95 | |||
| SP3; SPI1 | |||
| 40. | CDKAL1 | hsa-miR-370-5p; hsa-miR-873-5p.1; hsa-let-7b; hsa-let-7c; hsa-let-7d; hsa-let-7e; hsa-miR-145; hsa-miR-25; hsa-miR-301a; hsa-miR-301b; hsa-miR-451; hsa-miR-454; hsa-miR-495; hsa-miR-517b; hsa-miR-519a; hsa-miR-576-3p; hsa-miR-613; hsa-miR-616; hsa-miR-620; hsa-miR-650; hsa-miR-665; hsa-miR-766; hsa-miR-767-5p; hsa-miR-770-5p; hsa-miR-92a; hsa-miR-92b; hsa-miR-944 | CUX1; POU3F2 |
| 41. | TNF | hsa-miR-19a-3p; hsa-miR-203a-3p; hsa-miR-187-3p; hsa-miR-130a-3p; hsa-miR-143-3p; hsa-miR-130a;hsa-miR-130b; hsa-miR-149; hsa-miR-187 | AHR; ARNT; ATF1; ATF2; CEBPB; CEBPD; CREB1; EBF1; EGR1; EGR4; ELK1; ETS1; ETV4; FOS; IKBKB; IRF5 |
| JUN; NFAT5; NFATC1; NFATC2; NFATC3; NFATC4; NFE2L1; NFKB1; NFKB2; POU2F1; RELA; SMAD6; SMAD7; SP1; SP3; SPI1; STAT1; STAT2; STAT3; STAT4; STAT5A; STAT5B; STAT6; TBP; TFAP2A; TP53 | |||
| hsa-miR-19a; hsa-miR-296-3p; hsa-miR-409-5p; hsa-miR-454; hsa-miR-516a-5p; hsa-miR-516b; hsa-miR-519b-3p; hsa-miR-542-3p; hsa-miR-581; hsa-miR-592; hsa-miR-599; hsa-miR-654-3p; hsa-miR-770-5p; hsa-miR-875-3p; hsa-miR-875-5p; hsa-miR-939; hsa-miR-17; hsa-miR-9; hsa-miR-31 | |||
| 42. | TNXB | hsa-miR-30a-5p; hsa-miR-30b-5p; hsa-miR-30c-5p; hsa-miR-30d-5p; hsa-miR-30e-5p; hsa-miR-137; hsa-miR-146b-3p; hsa-miR-149; hsa-miR-152; hsa-miR-30a; hsa-miR-30a-5p; hsa-miR-30b; hsa-miR-30c; hsa-miR-30d; hsa-miR-30e; hsa-miR-372; hsa-miR-483-3p; hsa-miR-486-5p; hsa-miR-504; hsa-miR-512-3p; hsa-miR-638; hsa-miR-875-5p; hsa-miR-892b; hsa-miR-942 | ARNT; CTCF; E2F1; E2F2; E2F3; E2F4; E2F5; E2F6; E2F7; FOS; FOSB; FOSL1; HNF4A; JUN; JUNB; JUND; MAX; MEIS1; MYC; NFKB1; NR2F1; NR3C1; PAX2; PAX5; PPARG; RFX1; SREBF1; SREBF2; TFAP2A; TFAP2C; TGIF1; USF1; XBP1; YY1 |
| 43. | TRAF3IP2 | hsa-miR-3064-5p; hsa-miR-6504-5p; hsa-miR-147; hsa-miR-191; hsa-miR-30b; hsa-miR-30c; hsa-miR-342-5p; hsa-miR-512-3p; hsa-miR-548d-3p; hsa-miR-609; hsa-miR-637; hsa-miR-665; hsa-miR-765; hsa-miR-887; hsa-miR-935 | CUX1; FOXD3; FOXF2; FOXO4; IKBKB; IKBKG; |
| MAX; MXI1::CLEC5A; NKX2-2; NKX3-1; NR3C1; POU2F1; POU2F2; POU3F1; POU3F2; POU3F3; POU5F1; SRY; TCF3; USF1; ZEB1 | |||
| 44. | CCR6 | hsa-miR-518a-3p; hsa-miR-150-5p | CTCF |
| 45. | IL6 | hsa-let-7a-5p; hsa-miR-203a-3p; hsa-miR-142-3p; hsa-miR-26a-5p; hsa-miR-365a-3p; hsa-miR-107; hsa-let-7c-5p; hsa-miR-149-5p; hsa-miR-223-3p | AR; ATF1; CEBPA; CEBPB; CEBPD; CREB1; CTCF; EGR1; FOS; IRF1; IRF5; JUN; MYC; NFE2; NFIC; NFKB1; NFKB2; PBX1; PPARG; RARA; REL’ RELA; RREB1; STAT3; STAT5A; TP53 USF1; ZBTB16 |
| 46. | LYNX1 | hsa-miR-491-5p; hsa-miR-214; hsa-miR-324-3p; hsa-miR-324-5p; hsa-miR-330-5p; hsa-miR-370; hsa-miR-423-5p; hsa-miR-432; hsa-miR-511;hsa-miR-516a-5p; hsa-miR-526b; hsa-miR-608; hsa-miR-612; hsa-miR-637; hsa-miR-940; hsa-let-7a; hsa-let-7b; hsa-let-7c; hsa-let-7e; hsa-let-7f; hsa-let-7g; hsa-let-7i; hsa-miR-137; hsa-miR-144; hsa-miR-149; hsa-miR-153; hsa-miR-217; hsa-miR-338-5p; hsa-miR-365; | CTCF |
| hsa-miR-371-5p; hsa-miR-376a; hsa-miR-376b; hsa-miR-383; hsa-miR-548b-5p; hsa-miR-568; hsa-miR-574-3p; hsa-miR-587; hsa-miR-589; hsa-miR-655; hsa-miR-760; hsa-miR-98; hsa-miR-301b; hsa-miR-148a | |||
| hsa-miR-152; hsa-miR-519c; hsa-miR-301a | |||
| 47. | TNFSF8 | hsa-miR-24-3p; hsa-miR-146b-3p; hsa-miR-200b; hsa-miR-429; hsa-miR-525-3p; hsa-miR-626; hsa-miR-768-5p; hsa-miR-885-5p | NIL |
| 48. | TNFRSF1A | hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-22; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-558 | DAXX; EP300; IKBKB; IKBKG; JUN; STAT1 |
| 49. | VDR | hsa-miR-125b-5p; hsa-let-7a-5p; hsa-miR-27b-3p; hsa-miR-124; hsa-miR-125b; hsa-miR-506; hsa-miR-544 | BACH2; CREBBP; CTCF |
| FOS; FOSB; FOSL1; GTF2B; HMGN3;HNF4A; | |||
| HR; JUN; JUNB; JUND; KDM5A; LMO2; MAX; MED1; NCOA1; NCOA2; | |||
| NCOA6; NR0B2; NR1H2; | |||
| NRIP1; RXRA; RXRB; | |||
| RXRG; SMAD3; SNW1; | |||
| STAT1; TRIM24 | |||
| 50. | NOD2 | hsa-miR-122-5p; hsa-miR-122-3p; hsa-miR-495; hsa-miR-671-5p | MAX; MXI1::CLEC5A NFKB1; REL; SPI1; USF1 |
| 51. | STAT3 | hsa-miR-20b-5p; hsa-miR-337-3p; hsa-miR-21-5p; hsa-miR-92a-3p; hsa-miR-20a-5p; hsa-miR-124-3p; hsa-miR-130b-3p; hsa-miR-106a-5p; hsa-miR-106b-5p; hsa-miR-874-3p; hsa-miR-4516; hsa-miR-17-5p; hsa-miR-181a-5p; hsa-miR-1234-3p;hsa-miR-106a; hsa-miR-106b; hsa-miR-124; hsa-miR-125b; hsa-miR-130a; hsa-miR-17; hsa-miR-17-5p; hsa-miR-20a; hsa-miR-20b; hsa-miR-21; hsa-miR-372; hsa-miR-410; hsa-miR-495; hsa-miR-506; hsa-miR-519a; hsa-miR-519b-3p; hsa-miR-519c-3p; hsa-miR-519d; hsa-miR-665 hsa-miR-93 |
AR; ATF1; ATF2; ATF3; |
| ATF4; ATF5; ATF6; ATF7; BHLHE40; BRCA1; CEBPB; CREB1 | |||
| CREBBP; DAXX; EIF2AK2; EP300; FOXM1; GATA1; GATA2; GTF2I; HDAC1; HDAC2; HDAC3; HES1; | |||
| HIF1A; HNF1A; IRF9; JUN; KAT5; KHDRBS1; | |||
| MAX; MXI1::CLEC5A; | |||
| MYC; MYOD1; NCOA1; | |||
| NFKB1; NMI; NR3C1; | |||
| PIAS1; PIAS2; PIAS3; | |||
| PIAS4; PML; POU3F1; | |||
| PTMA; RARA; RELA; | |||
| STAT1; STAT4; STAT5A | |||
| STAT5B; TFAP2A; TP53 | |||
| USF1; ZNF148; ZNF467 | |||
| 52. | SLC9A3R1 | hsa-miR-24-3p; hsa-miR-146b-3p; hsa-miR-149; hsa-miR-200b; hsa-miR-200c; hsa-miR-24; hsa-miR-339-5p; hsa-miR-367; hsa-miR-532-5p; hsa-miR-548c-3p; hsa-miR-608; hsa-miR-632; hsa-miR-659; hsa-miR-663; hsa-miR-874 | CTCF; CTNNB1; E2F1; E2F2; E2F3; E2F4; E2F5; E2F6; E2F7; PPARG; SP1 |
| 53. | SOCS3 | hsa-miR-203a-3p; hsa-let-7f-5p; hsa-miR-19a-3p; hsa-miR-221-3p; hsa-miR-155-5p;hsa-miR-19a; hsa-miR-19b; hsa-miR-203; hsa-miR-218; hsa-miR-221; hsa-miR-30a; hsa-miR-30b | AHR; ARNT; E2F1; ESR1 |
| NFKB1; RELA; REST; STAT1; STAT2; STAT3 | |||
| hsa-miR-30e; hsa-miR-340; hsa-miR-561; hsa-miR-665; hsa-miR-765 | |||
| STAT4; STAT5A; STAT5B; STAT6; TCEB1; TCEB2; YY1 | |||
| 54. | BSG | hsa-miR-22-3p | EGR1; EGR2; MAX; MXI1::CLEC5A; MYC; TFAP2A; TFAP2C; USF1 |
| 55. | JUNB | hsa-miR-663a; hsa-miR-101; hsa-miR-199a-5p; hsa-miR-199b-3p; hsa-miR-30d; hsa-miR-30e; hsa-miR-328; hsa-miR-495; hsa-miR-526b; hsa-miR-566; hsa-miR-615-5p; hsa-miR-656; hsa-miR-663; hsa-miR-675; hsa-miR-744; hsa-miR-886-5p; hsa-miR-936 | ATF1; ATF2; ATF3; ATF4; ATF5; ATF6; ATF7; BATF; BCL6; BRCA1; CREB1; E2F1; ESR1; ETS2; FOS; FOSB; FOSL1; FOSL2; FOXO4; |
| JDP2; MAX; MXI1::CLEC5A, MYC | |||
| MZF1; NFE2L1; NFKB1 | |||
| SMAD3; SMAD4; SRF | |||
| TBP; TFAP2A; TFAP2C | |||
| TFAP4; USF1 | |||
| 56. | TGFB1 | hsa-miR-24-3p; hsa-miR-29b-3p; hsa-miR-144-3p; hsa-miR-633; hsa-miR-663a; hsa-miR-211-5p; hsa-miR-17-5p; hsa-miR-19b-3p; hsa-miR-93-5p; hsa-miR-324-3p; hsa-miR-122-5p; hsa-miR-130a-3p; hsa-miR-21; hsa-miR-24 | AR; CEBPA; CEBPB; CREB1; CTCF; DAXX; EGR1; EPAS1; FOS; GATA1; HIF1A; JUN; LMO2; MYC; MZF1; PAX5; PPARA; RARA; SMAD2; SMAD3; SMAD4; SP1; SP3; TP53; USF1; USF2; WT1; YY1 |
| 57. | RNF114 | hsa-miR-3064-5p; hsa-miR-6504-5p | EGR1; hsa-miR-124; hsa-miR-218; hsa-miR-492; hsa-miR-506; MAX; SP1; |
| USF1 | |||
| 58. | RPTOR | hsa-miR-99a; hsa-miR-100; hsa-miR-155-5p | CEBPA; DDIT3; RFX1; TLX2 |
| 59. | TGM1 | hsa-miR-130a; hsa-miR-130b; hsa-miR-142-3p; hsa-miR-148a; hsa-miR-148b; hsa-miR-149; hsa-miR-152; hsa-miR-301a; hsa-miR-301b; hsa-miR-345; hsa-miR-34a; hsa-miR-34c-5p; hsa-miR-361-3p; hsa-miR-378; hsa-miR-422a; hsa-miR-449a; hsa-miR-449b; hsa-miR-454; hsa-miR-502-5p; hsa-miR-508-5p; hsa-miR-558; hsa-miR-564; hsa-miR-617; hsa-miR-648; hsa-miR-920; hsa-miR-939 | AR; ESR1; HOXA7; RARA; RARB; RARG; TGIF1; TP53 |
| 60. | FABP5 | hsa-miR-144; hsa-miR-198; hsa-miR-203; hsa-miR-525-5p; hsa-miR-553; hsa-miR-562; hsa-miR-576-5p; hsa-miR-603; hsa-miR-616; hsa-miR-620 | CTCF; E2F1; MAX; MYC |
| 61. | TPPP | hsa-miR-1; hsa-miR-206 | Nil |
| 62. | WDR72 | hsa-miR-186; hsa-miR-576-5p; hsa-miR-599 | Nil |
| 63. | HMGCS2 | hsa-miR-490-5p | AR; CEBPA; NFIC; PPARA; RXRA; RXRB; RXRG; TFAP2A |
| 64. | TNNI2 | Nil | CTCF; CUX1; ELK1; POU2F1; RORA; SPI1; TFAP2A; TFAP2C |
| 65. | CNTNAP3B | Nil | HLF; STAT5A |
| 66. | ANKRD18A | hsa-miR-203; hsa-miR-518a-5p; hsa-miR-520g; hsa-miR-520h; hsa-miR-671-5p | Nil |
| 67. | ANKRD33B | Nil | EBF1; NFKB1; NFKB2; RELA; RELB; SPI1; TFAP2A; TFAP2C; USF1 |
| 68. | ERBB4 | hsa-miR-101; hsa-miR-106a; hsa-miR-125a-5p; hsa-miR-125b; hsa-miR-130a; hsa-miR-130b; hsa-miR-135a; hsa-miR-135b; hsa-miR-137; hsa-miR-144; hsa-miR-145; hsa-miR-146a; hsa-miR-146b-5p; hsa-miR-17; hsa-miR-184; hsa-miR-186; hsa-miR-199a-3p; hsa-miR-199b-3p; hsa-miR-19a; hsa-miR-19b; hsa-miR-200b; hsa-miR-200c; hsa-miR-205; hsa-miR-219-1-3p; hsa-miR-22; hsa-miR-221; hsa-miR-222; hsa-miR-23a; hsa-miR-23b | CEBPB; EP300; MEF2A; MEIS1; SMURF2; STAT5A; STAT5B; WWP1 |
| hsa-miR-26a; hsa-miR-26b; hsa-miR-300; hsa-miR-301; hsa-miR-301a; hsa-miR-301b; hsa-miR-302d; hsa-miR-323-3p; hsa-miR-330-3p; hsa-miR-335 | |||
| hsa-miR-339-5p; hsa-miR-340; hsa-miR-342-3p; hsa-miR-342-5p; hsa-miR-372; hsa-miR-377; hsa-miR-378 | |||
| hsa-miR-383; hsa-miR-410; hsa-miR-422a; hsa-miR-429; hsa-miR-432; hsa-miR-433; hsa-miR-454; hsa-miR-495; hsa-miR-507; hsa-miR-508-3p; hsa-miR-518a-5p; hsa-miR-519a; hsa-miR-519b-3p; hsa-miR-519c-3p; hsa-miR-519d; hsa-miR-520g; hsa-miR-520h; hsa-miR-527; hsa-miR-539; hsa-miR-548c-3p; hsa-miR-548d-3p; hsa-miR-571; hsa-miR-576-5p; hsa-miR-578; hsa-miR-579; hsa-miR-583; hsa-miR-584; hsa-miR-590-3p; hsa-miR-606; hsa-miR-653; hsa-miR-7; hsa-miR-876-5p; hsa-miR-93; hsa-miR-940; hsa-miR-944; hsa-miR-96 | |||
| 69. | TMEM132B | hsa-miR-10a; hsa-miR-10b; hsa-miR-137; hsa-miR-143; hsa-miR-148b; hsa-miR-152; hsa-miR-199a-5p; hsa-miR-19a; hsa-miR-19b; hsa-miR-218; hsa-miR-221; hsa-miR-222; hsa-miR-224; hsa-miR-297; hsa-miR-298; hsa-miR-338-3p; hsa-miR-377; hsa-miR-433; hsa-miR-548c-3p; hsa-miR-590-3p; hsa-miR-653; hsa-miR-766 | Nil |
| 70. | CNTNAP3 | hsa-miR-22; hsa-miR-26a; hsa-miR-26b; hsa-miR-9 | HLF; STAT5A |
| 71. | CNKSR2 | hsa-miR-144; hsa-miR-199b-5p; hsa-miR-21; hsa-miR-25; hsa-miR-28-3p; hsa-miR-29b; hsa-miR-30c; hsa-miR-32; hsa-miR-363; hsa-miR-374a; hsa-miR-374b; hsa-miR-450b-5p; hsa-miR-491-3p; hsa-miR-516b; hsa-miR-542-3p; hsa-miR-554; hsa-miR-627; hsa-miR-643; hsa-miR-942; hsa-miR-944 | BACH2; NFE2 |
| 72. | RAB3B | hsa-miR-194 | SMAD1; SMAD4; TLX2 |
| 73. | FREM2 | hsa-miR-142-3p; hsa-miR-142-5p; hsa-miR-147; hsa-miR-150; hsa-miR-200b; hsa-miR-200c; hsa-miR-24; hsa-miR-299-5p; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-30b; hsa-miR-32; hsa-miR-363; hsa-miR-367; | CTCF; EGR3; SP1; TBP; ZEB1 |
| hsa-miR-412; hsa-miR-429; hsa-miR-494; hsa-miR-509-3p; hsa-miR-544; | |||
| hsa-miR-548c-3p; hsa-miR-556-3p; hsa-miR-568; hsa-miR-580; hsa-miR-590-3p; hsa-miR-607; hsa-miR-628-3p | |||
| hsa-miR-633; hsa-miR-802; hsa-miR-885-5p; hsa-miR-9 | |||
| 74. | ACADL | hsa-miR-142-3p; hsa-miR-299-3p; hsa-miR-518a-5p; hsa-miR-641 | PPARD; RXRA; RXRB RXRG |
| 75. | CYP4A11 | hsa-miR-150 | AR; HNF4A; PPARA; PPARD; PPARG; RXRA; RXRB; RXRG |
| 76. | BTBD16 | hsa-miR-204; hsa-miR-211; hsa-miR-337-3p; hsa-miR-491-3p; hsa-miR-548c-3p; hsa-miR-599; hsa-miR-605; hsa-miR-625; hsa-miR-875-3p | CTCF; ELK1; TFAP2C; TLX2 |
| 77. | WFDC3 | hsa-miR-185; hsa-miR-28-5p; hsa-miR-29a; hsa-miR-29b; hsa-miR-29c; hsa-miR-329; hsa-miR-331-3p; hsa-miR-361-3p; hsa-miR-362-3p; hsa-miR-455-3p; hsa-miR-619; hsa-miR-657; hsa-miR-765; hsa-miR-875-3p; hsa-miR-923 | ATF1; ATF2; ATF3; ATF4; ATF5; ATF6; ATF7; CTCF; E2F1; ELK1; GABPA; JUN; NR3C1; RFX1; SP1; SPI1 |
| 78. | ACTC1 | hsa-miR-142-5p; hsa-miR-185; hsa-miR-195; hsa-miR-200b; hsa-miR-200c; hsa-miR-25; hsa-miR-30a; hsa-miR-30a-5p; hsa-miR-30b; hsa-miR-30c; hsa-miR-30d; hsa-miR-30e; hsa-miR-32; hsa-miR-324-3p; hsa-miR-340; hsa-miR-363; hsa-miR-367; hsa-miR-369-3p; hsa-miR-429; hsa-miR-495; hsa-miR-508-5p; hsa-miR-608; hsa-miR-7; hsa-miR-768-5p; hsa-miR-876-3p; hsa-miR-92a; hsa-miR-92b | BACH1; CUX1; IRF1; MEF2A; SRF; TFAP2A; TFAP2C; TGIF1; ZEB1 |
| 79. | LHCGR | hsa-miR-148b; hsa-miR-545 | NFIC; SP1; TFAP2A |
| 80. | FADS2 | hsa-let-7b | AR; CEBPA; CTCF; CUX1; DDIT3; HNF4A; MYC; PPARA; RXRA; RXRB; RXRG |
| 81. | GRIN2A | hsa-miR-101; hsa-miR-125a-5p; hsa-miR-125b; hsa-miR-137; hsa-miR-139-5p; hsa-miR-194; hsa-miR-19a; hsa-miR-19b; hsa-miR-206; hsa-miR-216b; hsa-miR-220b; hsa-miR-299-5p; hsa-miR-325; hsa-miR-329; hsa-miR-330-5p; hsa-miR-331-5p; hsa-miR-362-3p; hsa-miR-376a; hsa-miR-376b; hsa-miR-451; hsa-miR-454; hsa-miR-510; hsa-miR-519d; hsa-miR-520h; hsa-miR-525-5p; hsa-miR-574-3p; hsa-miR-576-5p; hsa-miR-577; hsa-miR-584; hsa-miR-593; hsa-miR-598; hsa-miR-603; hsa-miR-628-5p; hsa-miR-630; hsa-miR-656; hsa-miR-765 | AHR; ARNT; CTCF; PAX5; REST; USF1 |
| hsa-miR-767-5p; hsa-miR-9; hsa-miR-939 | |||
| 82. | SYT17 | hsa-miR-22; hsa-miR-297; hsa-miR-380; hsa-miR-574-5p; hsa-miR-633 | CTCF; MYC; PAX5; RREB1; TSC22D4 |
| 83. | RORC | hsa-let-7a; hsa-let-7b; hsa-let-7c; hsa-let-7e; hsa-let-7f; hsa-let-7g; hsa-let-7i | ARNT; ARNTL; CEBPA; CHD4; CLOCK; CTCF; FOXO4; LMO2; MAX; MXI1::CLEC5A; NCOA6 |
| hsa-miR-106b; hsa-miR-202; hsa-miR-205; hsa-miR-20a; hsa-miR-298; hsa-miR-485-5p; hsa-miR-519a; hsa-miR-519b-3p; hsa-miR-519c-3p; hsa-miR-593; hsa-miR-605; hsa-miR-608; hsa-miR-766; hsa-miR-93; hsa-miR-98 | NKX2-2; NPAS2; PPARG | ||
| SREBF1; SREBF2; TAL1 | |||
| TCF3; USF1; ZEB1 | |||
| 84. | CA1 | hsa-miR-944 | TBP |
| 85. | CHRM3 | hsa-miR-30c; hsa-miR-629 | TFAP2A |
| 86. | FPGT-TNNI3K | Nil | AHR; ARNT; E2F1; E2F2 |
| E2F3; E2F4; E2F5; E2F6 | |||
| E2F7; EGR1; YY1 | |||
| 87. | AGR3 | hsa-miR-32; hsa-miR-367; hsa-miR-448; hsa-miR-455-5p; hsa-miR-507; hsa-miR-548a-3p; hsa-miR-548c-3p; hsa-miR-557; hsa-miR-573; hsa-miR-656; hsa-miR-876-5p | Nil |
| 88. | SOHLH1 | hsa-miR-132; hsa-miR-220c; hsa-miR-484; hsa-miR-504; hsa-miR-516a-3p; hsa-miR-520a-5p; hsa-miR-525-5p; hsa-miR-600 | CTCF; USF1 |
| 89. | CYP1A2 | Nil | CUX1; NFIC;NKX6-1; TAL1; TCF4; USF1; USF2 |
| YY1 | |||
| 90. | CYP2W1 | hsa-miR-423-3p; hsa-miR-608; hsa-miR-637 | ZIC2 |
| 91. | BTC | hsa-miR-490-3p | CTCF; FOXC1; TFAP2A USF1 |
| 92. | DDC | Nil | AR; HNF1A; RORA |
Table 1: Psoriasis associated genes, miRNAs and transcription factors.
Construction of regulatory network (Cytoscape)
The regulatory network was constructed with 92 genes, 437 miRNAs and 285 TFs. Network was initiated by the Pubmed and Database Mining of 722 regulators (i.e., 285 TFs and 437 miRNAs) to interact with the 92 target genes in such a way to form 822 nodes and 2119 edges.
Identification of hub genes in regulatory network in topdown approach (Cytohubba)
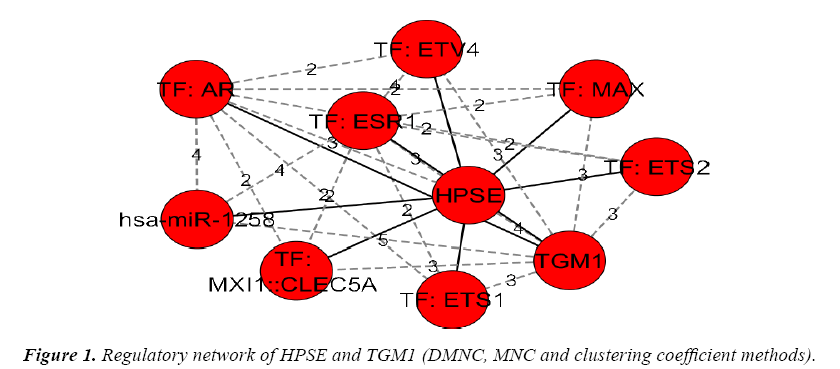
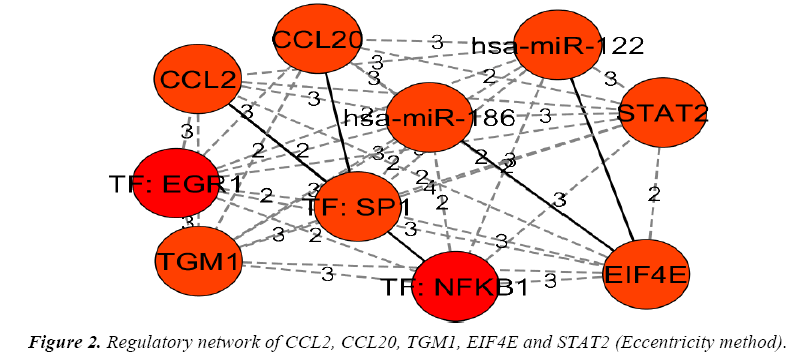
The genes and their regulators (Micro RNAs and transcription factors) were subjected to the analysis in cytohubba by various global based statistical methods like edge percolated component, bottleneck, eccentricity, closeness, radiality, betweenness and Stress along with local based statistical methods like maximal clique centrality, density of maximum neighborhood component, maximum neighborhood component and degree to identify their connectivity. Among the various methods of analysis in top down approach only a global based statistics of eccentricity method and the local based statistics of maximal clique centrality along with density of maximum neighborhood component and clustering coefficient methods in cytohubba resulted in obtaining a regulatory network of gene-miRNA-TFs in top-down approach. The details of regulatory network were given in Figures 1 and 2.
Implication of miRNAs in regulatory network
The implication of miRNAs in the regulatory network was analyzed on the basis of compatibility with respect to genemiRNA seed pairing and gene-miRNA-miRNA triplex with respect to nature of binding and the details were given in Table 2. In case of miRNAs implication in top-down approach hsamiR- 186-5p is highly compatible on the basis of seed pairing and triplex formation.
| S. No. | Genes | Micro RNAs | Binding Score in % (miRmap) |
Paired miRNA (Triplex RNA) |
Binding Energy in Kcal/mol. (Triplex RNA) |
Nature of Binding (Triplex RNA) |
|---|---|---|---|---|---|---|
| 1 | HPSE | hsa-miR-1258 | Nil | Nil | Nil | Nil |
| 2 | TGM1 | hsa-miR-1258 | Nil | Nil | Nil | Nil |
| hsa-miR-186 | Nil | Nil | Nil | Nil | ||
| hsa-miR-122 | Nil | Nil | Nil | Nil | ||
| 3 | CCL2 | hsa-miR-186 | Nil | Nil | Nil | Nil |
| hsa-miR-122 | Nil | Nil | Nil | Nil | ||
| 4 | CCL20 | hsa-miR-186 | 44.69 | Nil | Nil | Nil |
| hsa-miR-122 | Nil | Nil | Nil | Nil | ||
| 5 | EIF4E | hsa-miR-186 | 38.84 | hsa-miR-495 | -19.06 | miRNA self-complementarity |
| hsa-miR-122 | Nil | Nil | Nil | Nil | ||
| 6 | STAT2 | hsa-miR-186-5p | 47.13 | Nil | Nil | Nil |
| hsa-miR-122 | Nil | Nil | Nil | Nil |
Table 2: Implication of miRNA in regulatory network.
Pathway analysis
The obtained genes from Pubmed/DisGeNET/OMIM were subjected to pathway analysis in DAVID on the basis of P-value and Benjamini statistic and the result is given in Table 3. In case of pathway analysis, the genes associated with psoriasis follows the hierarchy of cytokine-cytokine receptor interaction, Jak-STAT signaling pathway, TNF signaling pathway and HIF- 1 signaling pathway.
| S. No. | Pathway | P value | Benjamini |
|---|---|---|---|
| 1. | Cytokine-cytokine receptor interaction | 8.60E-16 | 1.20E-13 |
| 2. | Jak-STAT signaling pathway | 2.40E-11 | 1.10E-09 |
| 3. | TNF signaling pathway | 1.00E-07 | 2.70E-06 |
| 4. | HIF-1 signaling pathway | 1.10E-04 | 1.10E-03 |
| 5. | FoxO signaling pathway | 5.00E-04 | 3.60E-03 |
| 6. | Osteoclast differentiation | 5.00E-04 | 3.60E-03 |