Research Article - Biomedical Research (2018) Volume 29, Issue 1
The role of insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) in endometrial carcinoma
Haixia Qin1,2, Danxia Chu2, Lijun Fan2, Yanmin Li2, Ya Xie2 and Ruixia Guo2*
1Department of Obstetrics and Gynecology, the First Affiliated Hospital of Xinxiang Medical University, Weihui, Henan, PR China
2Department of Obstetrics and Gynecology, the First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, PR China
- *Corresponding Author:
- Ruixia Guo
Department of Obstetrics and Gynecology
The First Affiliated Hospital of Zhengzhou University, PR China
Accepted date: September 7, 2017
DOI: 10.4066/biomedicalresearch.29-17-2121
Visit for more related articles at Biomedical ResearchAbstract
Objective: Insulin resistance is a high risk factor of Endometrial Carcinoma (EC). Insulin like Growth Factor Binding Protein-Related Protein 1 (IGFBP-rP1) could lead to insulin resistance due to its high affinity with insulin. The role of IGFBP-rP1 in EC is still unclear.
Method: The IGFBP-rP1 was overexpressed at the cellular level first, and then its effect on the growth of endometrial carcinoma cells was observed and its regulatory mechanism was investigated.
Results: Our study showed that IGFBP-rP1 upregulation decreased the proliferation of cells and the proportion of S+G2/M phase, but increased the apoptosis of human endometrial cancer cells HEC-1A. The expression of p-ERK1/2 was inhibited, while the concentration of rhIGFBP-rP1 increased. Nevertheless, IGFBP-rP1 was highly expressed in EC tissues. Further examination revealed that the DNA hypomethylation in the promoter region of IGFBP-rP1 was correlated with the high mRNA and protein expression levels of IGFBP-rP1 in EC tissues.
Conclusion: These results indicate that the DNA hypomethylation of IGFBP-rP1 can be used to predict high risk of EC. Further study may be required to confirm the predictor role of the methylation of IGFBP-rP1 in EC.
Keywords
Endometrial carcinoma, IGFBP-rP1, Methylation, ERK1/2
Introduction
Endometrial Carcinoma (EC) is one of the three most malignant tumors in female reproductive duct, accounting for 7% of female malignant tumor. According to the National Cancer Institute (NCI), the incidence of EC is becoming the highest in the malignant tumors of female reproductive duct [1]. Epidemiological studies have shown that metabolic syndrome, which is manifested as obesity, hypertension and diabetes, is a high risk factor to EC [2]. These high risk factors have a common pathophysiological basis, i.e. insulin resistance and the following hyperinsulinemia [3].
IGFBP-rP1 (Insulin-like Growth Factor Binding Proteinrelated Protein 1) is one type of proteins in IGFBPs (insulinlike growth factor binding protein system) which have relatively low binding with IGF (Insulin-like Growth Factor) [4]. Recent studies have shown that IGFBP-rP1 is not only involved in the formation of insulin resistance, but also plays a role in cell growth, aging and tumor formation. It is reported that IGFBP-rP1 can inhibit the growth of tumor cells such as breast cancer, lung cancer, malignant melanoma and colon cancer, and promote the apoptosis of tumor cells [5-8]. The use of IGFBP-rP1 in the transplant model of mice can slow tumor growth and inhibit tumor metastasis [7,9]. IGFBP-rP1 can also alter chemotherapeutic drug sensitivity [6,10]. Furthermore, IGFBP-rP1 exhibited low expression in breast cancer [11], colon cancer [12], prostate cancer [13] and other tumor tissue, indicating that IGFBP-rP1 might play a role as a tumor suppressor gene. Many studies show that IGFBP-rP1 inhibiting tumor might be related to the regulation of BRAF/MEK/ERK pathway. However, the role of IGFBP-rP1 in EC is still unclear. Previous studies have shown that low levels of IGFBP-rP1 and hyperinsulinemia in serum of patients with EC are risk factors for the formation of EC [14], and an improvement in insulin resistance or metabolic abnormalities can reduce the risk of EC. Therefore, IGFBP-rP1 may play a role in the formation of EC, similar to its role in other tumors, implying a potential therapeutic value for the anti-tumor treatment in EC.
In this study, we up-regulated IGFBP-rP1 at the cellular level first, and then observed its effect on the growth of endometrial carcinoma cells and investigated its regulatory mechanism. Finally, we confirmed the effect of IGFBP-rP1 on the growth regulation of EC on the tissue level.
Methods
Cell lines, transfection, plasmids and reagents
The human endometrial cancer cells HEC-1A were obtained from People’s Hospital of Beijing University, China. The cells were cultured in Dulbecco’s modified eagle medium (DMEM; Solarbio, China), supplemented with 10% fetal bovine serum and incubated under 5% CO2 at 37°C.
The plasmids contained the whole IGFBP-rP1 coding region (pEX-2-IGFBP7) and the negative control sequences (pEX-2- Empty) were designed and synthesized by Shanghai Jima Pharmaceutical Technology Company.
Cells were plated in a 6-well plate culture flask with 3 × 105 cells per well. After 20 h, Lipo2000 (Invitrogen, USA) was used as a transfection reagent according to the manufacturer’s instructions. The cells transferred with pEX-2-IGFBP7 or pEX-2-Empty were named as the experiment and negative control group, respectively. After 24 h from transfection, the experiment group, negative control group and their parental were collected to undergo cell proliferation, cell cycle and cell apoptosis analysis, respectively. After 36 h or 72 h from transfection, the three groups were collected to perform RNA or protein extraction, respectively.
RNA extraction and real-time PCR
Total RNA was isolated from cells and tissues using the TRIzol reagent (Cwbiotech, China) according to the manufacturer’s protocol. cDNA was synthesized by First Strand cDNA Synthesis Kit (Cwbiotech, China) according to the description of instruction. Real-time PCR was performed with SYBR Premix Ex TaqTM (TOYOBO, Japan) as the manufacturer’s protocol. The following protocol was used for real-time PCR amplification: preincubated at 95°C for 60 s followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. All the primers used in real-time PCR were described as follows: IGFBP-rP1 (forward primer 5’-CAGGTGTACTTGAGCTGTGAGG-3’, reverse primer 5’-CATGTAAGGCATCAACCACTGT-3’); β- actin (forward primer 5’-TGACGTGGACATCCGCAAAG-3’, reverse primer: 5’-CAGGAAGGTGGACAGCGAGG -3’). Data were analysed by the 2-DDCt method.
Western blot
This study used antibodies description as follows: IGFBP-rP1 (Abcam), ERK1/2 (Cell Signaling Technology), p-ERK1/2 (Cell Signaling Technology), GAPDH (Good Here,). Western blot was performed as literature described.
Cell proliferation, cell cycle and cell apoptosis assay
For cell proliferation assay, cells were collected and resuspended with Dulbecco’s modified eagle medium, seeded at 8 × 103 cells/well in 96-well plates and cultured in 100 μl Dulbecco’s modified eagle medium. The 10 μl cell counting kit-8 (CCK-8) was added to each well after being incubated for the indicated times (12, 24, 48 and 72 h) and then the samples were incubated at 37°C for 2 h and read by 96-well plate reader at 450 nm. All groups were done in triplicate wells.
For cell cycle analysis, cells were collected and fixed in 70% ethanol overnight at 4°C and then treated with the concentration of 10 μg/μl RNAase 2.5 μl for 30 min before with 50 μg/μl propidium iodide and analysed by Fluorescence Activated Cell Sorting (FACS).
Cell apoptosis examination was performed using an annexinVFITC apoptosis detection kit (Southern Biotech) according to the manufacturer’s instruction and then analysed by FACS.
Patients and samples
This study was approved by the Ethics Committee of the first affiliated hospital of Zhengzhou university and followed the Helsinki Declaration. We gathered 45 endometrioid adenocarcinoma, 30 endometrial dysplasia and 30 endometrial simple hyperplasia cases at the first affiliated hospital of Zhengzhou university (Zhengzhou, Henan, China) from October 2013 to May 2014. All cases were selected based on the following criteria: patients had pathologically confirmed diagnosis of EC or not, samples were stored at the pathology department of this hospital; patients were excluded if they underwent neoadjuvant chemotherapy or radiotherapy.
Immunohistochemistry (IHC) and evaluation
The rabbit polyantibody of IGFBP-rP1 was used in IHC to be primary antibody. The IHC was undergoing as the manufacturer’s protocol (Cwbiotech, China).
Using the staining results of cytoplasm or cell membrane as the criterion, five high-magnification fields were randomly observed by reference to the semi-quantitative fractionation of positive cells such as Fromowitz, and 100 cells were counted for each field of view.
According to the percentage of positive cells, marking was carried out: <5% for 0 point, 5% to 25% for 1 point, 26% to 50% for 2 points, 51% to 75% for 3 points and >75% for 4 points. According to the positive cell staining intensity, the score was: no colour for 0 point, light yellow for 1 point, brown-yellow for 2 points, brown for 3 points.
According to the total points obtained, the results were categorised as follows: 0-1 point indicates negative (-), 2~3 points indicate weak positive (+), 4~5 points indicate moderate positive (++), 6~7 points indicate strong positive (+++).
DNA methylation assay
According to the manufacturer’s protocol, Genomic DNA was isolated from tissues using the Genome DNA extracts kit (Cwbiotech, China) and the DNA methylation was detected by EZ DNA Methylation-gold kit (ZYMO Research, USA). All the primers and annealing temperature of its primer used in PCR amplification were described in Table 1. The PCR products were performed to agarose electrophoresis.
| Primer | Sequence | Tm (t/°C) | Product (bp) |
|---|---|---|---|
| Promoter region | |||
| Methylation | F: 5’-AGAAATTAGAGGGTGGAAGAGTCGT-3’ R: 5’-CTACTAACGTCGAAAAATAAACGAA-3’ |
54 | 173 |
| Nonmethylation | F:5’-AGAAATTAGAGGGTGGAAGAGTTG-3’ R: 5’-CTACTAACATCAAAAAATAAACAAA-3’ |
50 | 173 |
| First exon region | |||
| Methylation | F: 5’-TTGGGCGAGATTCGCGACGC-3’ R: 5’-GACCCTCTAACTAACGACGCG-3’ |
60 |
273 |
| Nonmethylation | F: 5’-TTGTTGGGTGAGATTTGTGATGT-3’ R: 5’-TCAACCCTCTAACTAACAACACA-3’ |
54 | 278 |
Table 1. The primers of the IGFBP-rP1 methylation.
Statistical analyses
Statistical analysis was carried out by means of SPSS (version 17.0; SPSS Inc., Cary, NC, USA). Chi-squared and Student’s ttest were used to determine statistical significance at p<0.05. The correlation between the DNA methylation and the expression of IGFBP-rP1 in EC was analysed by Spearman coefficients. All data were expressed as and all experiments described above were performed in triplicate.
Results
Efficient up regulation of IGFBP-rP1 in HEC-1A cells
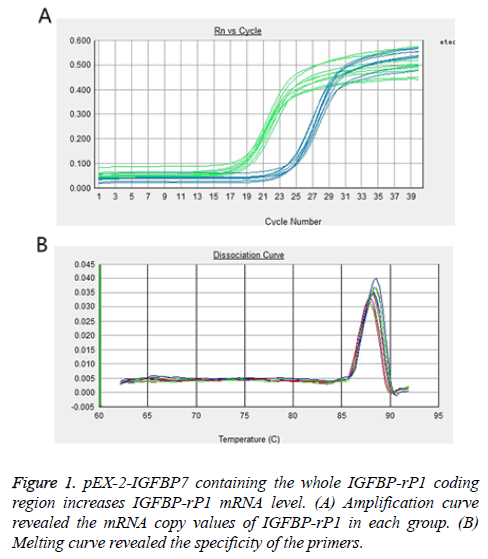
Real-time PCR and Western blot were used to analyse IGFBP-rP1 gene and protein expression in experiment group, negative control group and their parental, respectively. As shown in Figure 1, the mRNA copy values of IGFBP-rP1 in experiment group, negative control group and their parental were 2.699 ± 0.293, 1.296 ± 0.169 and 1.031 ± 0.301, respectively, which shows that the gene expression of IGFBP-rP1 in experiment group was significantly higher than that in negative control group (P=0.001), whereas no apparent change was observed in negative control group and their parental (P=0.261).
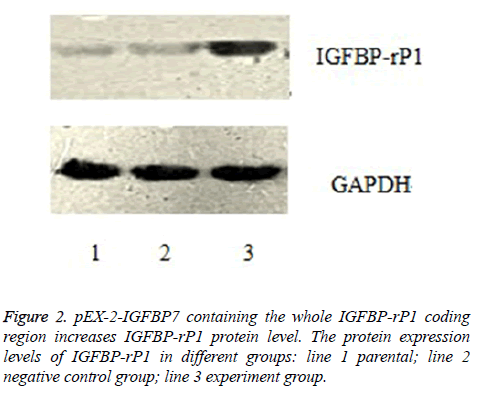
As shown in Figure 2, the relative protein expression levels of IGFBP-rP1 in experiment group, negative control group and their parental were 1.126 ± 0.074, 0.889 ± 0.041 and 0.884 ± 0.042, respectively. They were shown a significantly high lever in experiment group than in negative control group (P<0.01), whereas no apparent change was observed in negative control group and their parental (P>0.05).
IGFBP-rP1 upregulation decreases the cell proliferation, the proportion of cells in the S+G2/M phase; but increases the cell apoptosis
After 24 h, 48 h and 72 h from transfection, the cellular proliferation inhibition rate in experiment group was 0.373 ± 0.054, 0.399 ± 0.047 and 0.380 ± 0.053, respectively. Correspondingly, the cellular proliferation inhibition rate in negative control group was 0.036 ± 0.006, 0.040 ± 0.005, and 0.0334 ± 0.006, while it was 0.027 ± 0.003, 0.032 ± 0.002 and 0.024 ± 0.002 in parental group. The results showed that the cell proliferation is significantly decreased after IGFBP-rP1 upregulation (P<0.01). However, there was no statistically significant difference between negative control group and parental group (P>0.05).
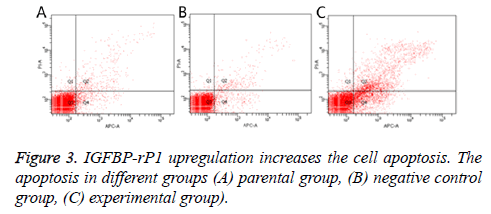
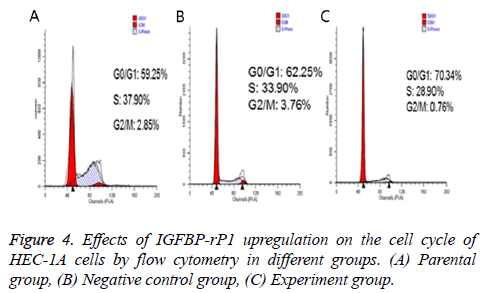
Twenty-four hours after transfection, the apoptotic rate was 20.667% ± 2.055% in the experimental group, 3.967% ± 0.351% in the negative control group and 4.967% ± 0.252% in the parental group. The experimental group has statistically significant difference in comparison with the negative control group and parental group (P<0.01), but there was no statistically significant difference between the control group and the parental group (P>0.05) (Figure 3). The ratio of S +G2/M phase cells in the experimental group was 30.980% ± 1.461%, which was significantly lower than that of the negative control group and the parental group (38.797% ± 2.419% and 37.800% ± 0.350%, respectively; P<0.01) There was no statistically significant difference in the cell cycle between the negative control group and the parental group (P>0.05) (Figure 4).
Effects of different concentrations of rhIGFBP-rP1 on proliferation of HEC-1A cells and ERK pathway activation in EC
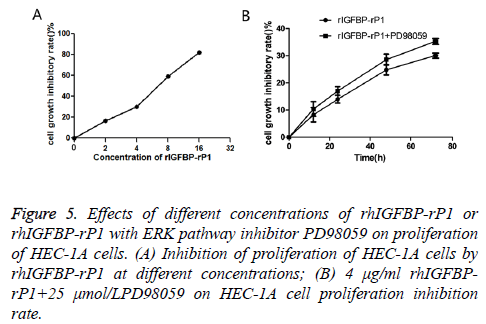
In the experimental groups, HEC-1A cells were incubated with rhIGFBP-rP1 at concentrations of 2 μg/ml, 4 μg/ml, 8 μg/ml and 16 μg/ml, respectively. After 72 h, compared with the control group, the OD value of each experimental group decreased (the higher the rhIGFBP-rP1 concentration, the more significant the decrease) and cell proliferation was inhibited. Inhibition rate=(ODcontrol group-ODexperimental group)/ODcontrol group × 100.
As shown in Figure 5A, the inhibitory rates of rhIGFBP-rP1 on HEC-1A were (16.58 ± 1.72%), (30.04 ± 0.94%), (58.74 ± 1.29) and (81.89 ± 0.87%) (P<0.05) for rhIGFBP-rP1 concentration of 5 μg/ml, 4 μg/ml, 8 μg/ml and 16 μg/ ml, respectively. Based on the above data, a statistical analysis using SPSS17.0 obtained IC50=6.39 μg/ml. The inhibitory effect of 4 μg/ml rIGFBP-rP1 on HEC-1A proliferation was more and more obvious with the prolongation of time.
Figure 5: Effects of different concentrations of rhIGFBP-rP1 or rhIGFBP-rP1 with ERK pathway inhibitor PD98059 on proliferation of HEC-1A cells. (A) Inhibition of proliferation of HEC-1A cells by rhIGFBP-rP1 at different concentrations; (B) 4 μg/ml rhIGFBPrP1+ 25 μmol/LPD98059 on HEC-1A cell proliferation inhibition rate.
As shown in Figure 5B, the inhibitory rates were (8.25 ± 2.65%), (13.98 ± 1.41%), (24.82 ± 1.90%) and (30.04 ± 0.94%) (P<0.05) for proliferation after 6, 12, 24, 48 and 72 h, respectively. With combination of rhIGFBP-rP1 (4 μg/ml) and ERK pathway inhibitor PD98059 (25 umol/L), the inhibitory rate of cell proliferation increased to (10.04 ± 2.71%), (17.02 ± 1.58%), (28.59 ± 2.04%) and (35.29 ± 1.12%) (P<0.05) for the aforementioned proliferation time.
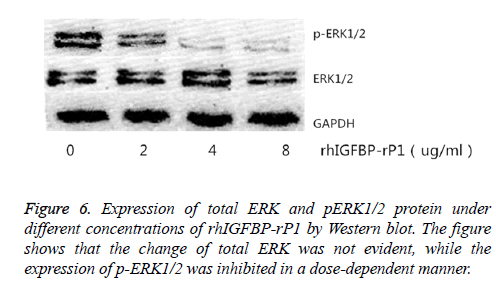
The change in the expression of total ERK1/2 and p-ERK1/2 protein in the cells was observed by using Western blot after 24 h of treatment with different concentrations of rhIGFBP-rP1 (0 μg/ml, 2 μg/ml, 4 μg/ml, 8 μg/ml). Western blot test showed that the change of total ERK was not evident, while the expression of p-ERK1/2 was inhibited in a dose-dependent manner, as shown in Figure 6.
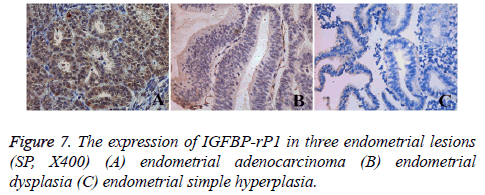
The expression of IGFBP-rP1 in three endometrial lesions
Tissue immunohistochemistry showed that IGFBP-rP1 was expressed in the cytoplasm and membrane (Figure 7). Overall, a large number of brown stained cells were observed in endometrioid adenocarcinoma tissue, and they were deep stained. There were also many brown stained cells in atypical hyperplasia tissues of endometrium, and the staining was darker. No obvious brown stain was seen in endometrial simple hyperplasia tissue. The positive expression rate of IGFBP-rP1 in endometrial adenocarcinoma (30/45, 66.7%) was significantly higher than that in endometrial dysplasia (5/30, 16.7%) (χ2=18.080, P=0.00<0.016) and simple endometrial hyperplasia (7/30, 23.3%) (χ2=13.522, P=0.00<0.016), but there was no statistically significant difference between atypical hyperplasia and simple hyperplasia (χ2=0.417, P=0.519>0.016).
RT-PCR results showed that the relative expression of IGFBP-r P1 m RNA in endometrioid adenocarcinoma was significantly higher than that in endometrial dysplasia (t=5.17, P<0.01) and simple endometrial hyperplasia (t=6.56, P<0.01), but there was no significant difference between the first two groups (t=1.33, P>0.05, Table 2).
| Groups | ΔCT | ΔΔCT | 2-ΔΔCT |
|---|---|---|---|
| Endometrial simple hyperplasia | 6.132 ± 2.390 | 0.000 ± 2.390 | 1 |
| Endometrial dysplasia | 6.432 ± 1.170 | 0.300 ± 1.170 | 0.81 |
| Endometrial adenocarcinoma | 2.317 ± 4.372 | -3.815 ± 4.372 | 14.07 |
Table 2. The mRNA levels of IGFBP-rP1 in three endometrial lesions.
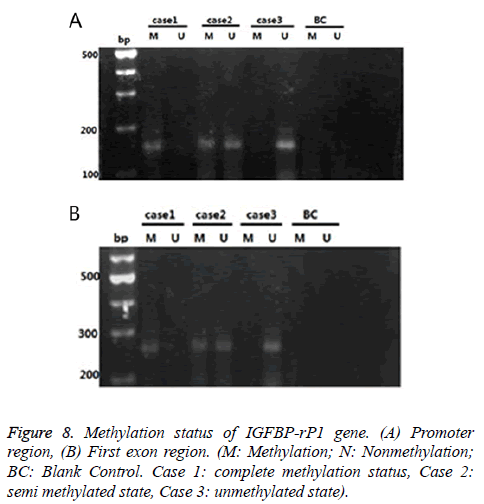
IGFBP-rP1 gene in the endometrial adenocarcinoma exhibited a methylation status mainly in promoter region, and showed a low methylation status
There were three cases for the methylation status of the IGFBP-rP1 gene in the tissue: complete methylation, unmethylation and halomethylation (Figure 8), where complete methylation and halomethylation were recorded in methylation.
The methylation rate (15/45) of the IGFBP-rP1 gene in the promoter region of endometrial adenocarcinoma was significantly lower than that in the untypical endometrial dysplasia tissues (20/30) (χ2=8.036, P=0.005<0.016) and that in the endometrial dysplasia (21/30) (χ2=9.696, P=0.002<0.016). There was no significant difference in the methylation rate of IGFBP-rP1 gene in the promoter region between the endometrial hyperplasia tissues and the simple hyperplasia tissues (χ2=0.077, P=0.781>0.016).
The methylation rate (5/45) of the IGFBP-rP1 gene in the first exon region of endometrial adenocarcinoma had no significant difference compared with those in the untypical endometrial dysplasia (7/30) and simple hyperplasia (8/30) (χ2=3.325, P=0.190>0.05).
The methylation rate (15/45) of the IGFBP-rP1 gene in the promoter region of endometrial adenocarcinoma was higher than that in the first exon region (5/45) (χ2=6.429, P=0.011<0.05).
IGFBP-rP1 gene was highly expressed in endometrial adenocarcinoma and was negatively correlated with its methylation
In the promoter region, the mRNA expression level of IGFBP-rP1 gene in methylated endometrial adenocarcinoma was significantly higher than that in non-adimal endometrial adenocarcinoma (t=4.137, P<0.05). The mRNA expression level of endometrial adenocarcinoma in the first exon region was not significantly different from that in the endometrial adenocarcinoma (t=2.123, P>0.05).
In the promoter region, 13 cases showed positive protein expression in 15 cases of endometrial adenocarcinoma with IGFBP-rP1 gene methylation, which was significantly higher than that of non-methylated endometrial adenocarcinoma. The methylation degree of IGFBP-rP1 was negatively correlated with protein expression (χ2=4.050, P<0.05, r=-0.287). In the first exon region, there was no significant difference in the protein expression between the endometrial adenocarcinoma and the non-endometrial adenocarcinoma (t=2.123, P>0.05).
Discussion
In recent years, the incidence of endometrial cancer worldwide increased year by year. It is well known that the long-term stimulation of high levels of estrogen without progesterone antagonism is closely related to the occurrence of endometrial cancer, but the mechanism on the pathogenesis of endometrial cancer has not been yet fully understood. Recent studies have found that obesity, hypertension, diabetes, dyslipidemia, and other high risk factors, which collectively constitute metabolic syndrome [15], have become high risk factors for a variety of tumors [16-18]. In particular, the occurrence of endometrial cancer is also closely related to these metabolic disorders [17,18]. The obesity, hypertension and diabetes all have pathophysiological characteristics manifested as insulin resistance and the subsequent hyperinsulinemia [15]. Some recent studies have also found that elevated insulin levels, independent of estradiol, age and hormone replacement treatment, are closely related with endometrial cancer [19].
Insulin-like growth factor system is composed of ligand, cell surface receptor, insulin receptor, IGF binding protein and IGFBP protease composition, and it is the focus in studies of insulin resistance. IGFBP-rP1 is an important member of the IGFBPs family and has been extensively studied because of its low binding to insulin growth factors. Many studies have shown that IGFBP-rP1 plays a role in tumor suppressor genes in various tumors, such as breast cancer [8,20-22], hepatocellular carcinoma [23] and lung cancer [24]; however, it can be expressed as a carcinogenic effect on glioma [25]. Previous investigations showed that IGFBP-rP1 also plays an important role in the neuroendocrine differentiation for certain type of tumor [26]. It has been largely accepted that tumor with neuroendocrine differentiation is one of the most important prognostic factors [27-29]. In the view of the neuroendocrine differentiation, investigations of IGFBP-rP1 in EC have greater significance.
In order to study the role of IGFBP-rP1 in endometrial carcinoma, IGFBP-rP1 cDNA was transfected into endometrial carcinoma cell line HEC-1A with low expression of endogenous IGFBP-rPl. Realtime-PCR and Westem blot confirmed increase in IGFBP-rP1 expression, and then a series of analyses were carried out for the biological function of the IGFBP-rP1 in endometrial cancer cells. The expression of IGFBP-rP1 in endometrial carcinoma cells was significantly increased, the apoptosis rate of endometrial carcinoma cells was significantly decreased, and the apoptosis rate of G0/G1 cells was significantly increased. Such effects are similar to those observed in prostate cancer, breast cancer and lung cancer, etc. [11,13,30-32]. This suggests that IGFBP-rP1 is most likely to play a role in tumor suppression in endometrial cancer cells. In addition, we investigated the mechanism of IGF BP-rP1 suppressing the growth of endometrial carcinoma cells, and found that IGFBP-rP1 reduced the level of phosphorylated ERK1/2 protein by mediating the MEK/ERK signal transduction pathway after insulin receptor, thereby inhibiting endometrial cancer cell proliferation. This function can be enhanced by MEK/ERK signaling pathway inhibitor PD98059. In other words, IGFBP-rP1 can more effectively inhibit endometrial cancer cells HEC-1A with help from the MER/ERK signal pathway inhibitor PD98059.
However, when the inhibitory effect of IGFBP-rP1 was examined using histology, the expression rate of IGFBP-rP1 in endometrioid adenocarcinoma was significantly higher than that in non-typical endometrial dysplasia and simple endometrial dysplasia. These results suggesting that IGFBPrP1 seems to play a cancer-causing factor on tissue level, which contradicts the results of cell experiments.
The contradictory results of IGFBP-rP1 in the endometrial cancer in vivo and in vitro suggest that the role of IGFBP-rP1 in endometrial cancer may be pre-transcriptional regulation of genes. Many recent studies have shown that IGFBP-rP1 methylation is correlated with tumor development and progression. IGFBP-rP1 gene hypermethylation status also appears in a variety of tumors such as laryngeal cancer [33], human melanoma cell line [34], cardia adenocarcinoma [35], gastric cancer cells [36], Barrett esophagus and esophageal squamous cell carcinoma [37], colon cancer [20] and breast cancer [38,39]. These studies have shown that IGFBP-rP1 hypermethylation state in the promoter region was correlated with its downregulation. In this study, we found that the methylation rate of IGFBP-rP1 gene in the promoter region of endometrial adenocarcinoma was higher than that in the first exon region, and its abnormal methylation was mainly in the promoter region. Further investigation has shown that the methylation rate of the IGFBP-rP1 gene in the endometrial adenocarcinoma group was significantly lower in the promoter region than in the non-typical endometrial dysplasia group and the simple endometrial hyperplasia group. The methylation rate of the two latter groups was not statistically significant, and the low methylation level of this gene in the endometrial adenocarcinoma group was negatively correlated with its high expression status. IGFBP-rP1 gene also showed low methylation levels in colorectal cancer tissues. Lin et al. [20] found that the methylation rate in the first exon region of IGFBP-rP1 gene was significantly lower in colorectal cancer tissues. There was a negative correlation between the low methylation level and the high expression of the gene in the normal mucosal tissue. The mRNA and protein expressions of IGFBP-rP1 gene in the endometrial adenocarcinoma and colorectal tissue were significantly higher, suggesting that the expression of this gene regulation occurs at the transcription level or before the molecular events, while DNA abnormal methylation occurs at this level of regulation. This shows the correlation between methylation and expression.
Abnormal methylation of DNA is a common molecular event that occurs in the early stages of human tumors and is a further refinement and development of the Knudson secondary strike mechanism [40]. Once methylation occurs, the transcriptional regulation of the corresponding gene is affected, which inhibits gene expression [41-46]. The present study suggests that the hypermethylation state in the promoter region affects the transcription of the gene, thereby influencing the expression of the gene. This is one of the mechanisms that inhibit the antitumor effect of IGFBP-rP1 gene, which is related to the occurrence and development of tumor. Since DNA methylation is a reversible chemical modification, DNA demethylation may restore the tumor suppressor gene activation and play an antitumor effect, which suggests that this principle can be used in clinical anti-tumor treatment, providing new solution to treat cancer.
In summary, this study first found that upregulation of IGFBPrP1 in endometrial cancer cells can inhibit cell proliferation by inhibiting MEK/ERK pathway. On the other hand, histological studies have shown that overexpression of IGFBP-rP1 was associated with malignancy of endometrial cancer. Further studies have found that the degree of methylation of IGFBPrP1 in endometrial carcinoma was reduced, and it was negatively correlated with its expression. These results show that the increase in the expression of mRNA and protein due to the reduction in methylation of IGFBP-rP1 gene promoter may be a predictor of the degree of malignancy of endometrial cancer. The occurrence of cancer is a complex process and thus in the future a further study of the regulation mechanism of IGFBP-rP1 expression may provide new ideas for clinical diagnosis and treatment of endometrial adenocarcinoma.
Acknowledgment
None
Grant Support
This work is supported by National Natural Science Foundation of China, Grant (No. 31670844); Henan University Science and Technology innovation team support program, Grant (No. 17IRTSTHN021).
Ethic Approval
This study was approved by the Ethics Committee of the first affiliated hospital of Zhengzhou university and followed the Helsinki Declaration.
References
- Kutsukake M, Ishihara R, Momose K, Isaka K, Itokazu O, Higuma C, Matsutani T, Matsuda A, Sasajima K, Hara T, Tamura K. Circulating IGF-binding protein 7 (IGFBP7) levels are elevated in patients with endometriosis or undergoing diabetic hemodialysis. Reprod Biol Endocrinol 2008; 6: 54.
- Esposito K, Chiodini P, Capuano A, Bellastella G, Maiorino MI. Metabolic syndrome and endometrial cancer: a meta-analysis. Endocrine 2014; 45: 28-36.
- Zhang Y, Liu Z, Yu X, Zhang X, Lu S, Chen X, Lü B. The association between metabolic abnormality and endometrial cancer: A large case-control study in China. Gynecol Oncol 2010; 117: 41-46.
- Hwa V, Oh Y, Rosenfeld RG. The insulin-like growth factor-binding protein (IGFBP) superfamily. Endocr Rev 1999; 20: 761-787.
- Ruan W, Xu E, Xu F, Ma Y, Deng H, Huang Q, Lv B, Hu H, Lin J, Cui J, Di M, Dong J, Lai M. IGFBP7 plays a potential tumor suppressor role in colorectal carcinogenesis. Cancer Biol Ther 2007; 6: 354-359.
- Okamura J, Huang Y, Moon D, Brait M, Chang X, Kim MS. Downregulation of insulin-like growth factor-binding protein 7 in cisplatin-resistant non-small cell lung cancer. Cancer Biol Ther 2012; 13: 148-155.
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 2008; 132: 363-374.
- Landberg G, Ostlund H, Nielsen NH, Roos G, Emdin S, Burger AM, Seth A. Downregulation of the potential suppressor gene IGFBP-rP1 in human breast cancer is associated with inactivation of the retinoblastoma protein, cyclin E overexpression and increased proliferation in estrogen receptor negative tumors. Oncogene 2001; 20: 3497-3505.
- Chen D, Siddiq A, Emdad L, Rajasekaran D, Gredler R, Shen XN, Santhekadur PK, Srivastava J, Robertson CL, Dmitriev I, Kashentseva EA, Curiel DT, Fisher PB, Sarkar D. Insulin-like growth factor-binding protein-7 (IGFBP7): a promising gene therapeutic for hepatocellular carcinoma (HCC). Mol Ther 2013; 21: 758-766.
- Tomimaru Y, Eguchi H, Wada H, Noda T, Murakami M, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M, Nagano H. Insulin-like growth factor-binding protein 7 alters the sensitivity to interferon-based anticancer therapy in hepatocellular carcinoma cells. Br J Cancer 2010; 102: 1483-1490.
- Smith P, Nicholson LJ, Syed N, Payne A, Hiller L, Garrone O, Occelli M, Gasco M, Crook T. Epigenetic inactivation implies independent functions for insulin-like growth factor binding protein (IGFBP)-related protein 1 and the related IGFBPL1 in inhibiting breast cancer phenotypes. Clin Cancer Res 2007; 13: 4061-4068.
- Ruan W, Zhu S, Wang H, Xu F, Deng H. IGFBP-rP1, a potential molecule associated with colon cancer differentiation. Mol Cancer 2010; 9: 281.
- Nousbeck J, Ishida-Yamamoto A, Bidder M, Fuchs D, Eckl K, Hennies HC, Sagiv N, Gat A, Gini M, Filip I, Matz H, Goldberg I, Enk CD, Sarig O, Meilik B, Aberdam D, Gilhar A, Sprecher E. IGFBP7 as a potential therapeutic target in Psoriasis. J Invest Dermatol 2011; 131: 1767-1770.
- Zhan Y, Wang J, Ma Y, Liu Z, Xu H, Lu S, Lu B. Serum insulin-like, growth factor binding protein-related protein 1 (IGFBP-rP1) and endometrial cancer risk in Chinese women. Int J Cancer 2013; 132: 411-416.
- DeFronzo RA, Ferrannini E. Insulin resistance. A multifaceted syndrome responsible for NIDDM, obesity, hypertension, dyslipidemia, and atherosclerotic cardiovascular disease. Diab Care 1991; 14: 173-194.
- Rose DP, Komninou D, Stephenson GD. Obesity, adipocytokines, and insulin resistance in breast cancer. Obes Rev 2004; 5: 153-165.
- Soliman PT, Wu D, Tortolero-Luna G, Schmeler KM, Slomovitz BM, Bray MS, Gershenson DM, Lu KH. Association between adiponectin, insulin resistance, and endometrial cancer. Cancer 2006; 106: 2376-2381.
- Djiogue S, Nwabo Kamdje AH, Vecchio L, Kipanyula MJ, Farahna M, Aldebasi Y, Seke Etet PF. Insulin resistance and cancer: the role of insulin and IGFs. Endocr Relat Cancer 2013; 20: 1-17.
- Hernandez AV, Pasupuleti V, Benites-Zapata VA, Thota P, Deshpande A, Perez-Lopez FR. Insulin resistance and endometrial cancer risk: a systematic review and meta-analysis. Eur J Cancer 2015; 51: 2747-2758.
- Lin J1, Lai M, Huang Q, Ma Y, Cui J. Methylation patterns of IGFBP7 in colon cancer cell lines are associated with levels of gene expression. J Pathol 2007; 212: 83-90.
- Benatar T, Yang W, Amemiya Y, Evdokimova V, Kahn H, Holloway C, Seth A. IGFBP7 reduces breast tumor growth by induction of senescence and apoptosis pathways. Breast Cancer Res Treat 2012; 133: 563-573.
- Burger AM, Zhang X, Li H, Ostrowski JL, Beatty B, Venanzoni M, Papas T, Seth A. Down-regulation of T1A12/mac25, a novel insulin-like growth factor binding protein related gene, is associated with disease progression in breast carcinomas. Oncogene 1998; 16: 2459-2467.
- Chen D, Yoo BK, Santhekadur PK, Gredler R, Bhutia SK, Das SK, Fuller C, Su ZZ, Fisher PB, Sarkar D. Insulin-like growth factor-binding protein-7 functions as a potential tumor suppressor in hepatocellular carcinoma. Clin Cancer Res 2011; 17: 6693-6701.
- Chen Y, Pacyna-Gengelbach M, Ye F, Knosel T, Lund P, Deutschmann N, Schluns K, Kotb WF, Sers C, Yasumoto H, Usui T, Petersen I. Insulin-like growth factor binding protein-related protein 1 (IGFBP-rP1) has potential tumour-suppressive activity in human lung cancer. J Pathol 2007; 211: 431-438.
- Jiang W, Xiang C, Cazacu S, Brodie C, Mikkelsen T. Insulin-like growth factor binding protein 7 mediates glioma cell growth and migration. Neoplasia 2008; 10: 1335-1342.
- Walker GE, Antoniono RJ, Ross HJ, Paisley TE, Oh Y. Neuroendocrine-like differentiation of non-small cell lung carcinoma cells: regulation by cAMP and the interaction of mac25/IGFBP-rP1 and 25.1. Oncogene 2006; 25: 1943-1954.
- Okubo Y, Motohashi O, Nakayama N, Nishimura K, Kasajima R, Miyagi Y, Shiozawa M, Yoshioka E, Suzuki M, Washimi K, Kawachi K, Nito M, Kameda Y, Yokose T. The clinicopathological significance of angiogenesis in hindgut neuroendocrine tumors obtained via an endoscopic procedure. Diagn Pathol 2016; 11: 128.
- Okubo Y, Nemoto T, Wakayama M, Tochigi N, Shinozaki M, Ishiwatari T, Aki K, Tsuchiya M, Aoyama H, Katsura K, Fujii T, Nishigami T, Yokose T, Ohkura Y, Shibuya K. Gangliocytic paraganglioma: a multi-institutional retrospective study in Japan. BMC Cancer 2015; 15: 269.
- Okubo Y, Yokose T, Motohashi O, Miyagi Y, Yoshioka E, Suzuki M, Washimi K, Kawachi K, Nito M, Nemoto T, Shibuya K, Kameda Y. Duodenal rare neuroendocrine tumor: clinicopathological characteristics of patients with gangliocytic paraganglioma. Gastroenterology Res Pract 2016; 2016: 5257312.
- Doverskog M, Tally M, Haggstrom L. Constitutive secretion of an endogenous insulin-like peptide binding protein with high affinity for insulin in Spodoptera frugiperda (Sf9) cell cultures. Biochem Biophys Res Commun 1999; 265: 674-679.
- Yamanaka Y, Wilson EM, Rosenfeld RG, Oh Y. Inhibition of insulin receptor activation by insulin-like growth factor binding proteins. J Biol Chem 1997; 272: 30729-30734.
- Berstein LM, Kvatchevskaya JO, Poroshina TE, Kovalenko IG, Tsyrlina EV, Zimarina TS, Ourmantcheeva AF, Ashrafian L, Thijssen JH. Insulin resistance, its consequences for the clinical course of the disease, and possibilities of correction in endometrial cancer. J Cancer Res Clin Oncol 2004; 130: 687-693.
- Wang JT. Study on methylation and protein expression of IGFBP-rP1 promoter in laryngeal squamous cell carcinoma. J Hebei Med Univ 2012: 1-32;
- Xie YN, Chen Hao, Zhou WQ, Cao YP, Tao Y, Zeng XS, Sun JF. Study on abnormal methylation of CpG Island in promoter region of human melanoma cell line IGFBP-7. Chinese J Dermatol 2012; 45: 714-717.
- Shen SP, Dong ZM, Guo W, Guo YL, Kuang G, Yang ZB. Methylation status of IGFBP7 gene in human gastric cardia adenocarcinoma. Chinese J Cancer Biother 2012; 19: 632-638.
- He DZ, Li JS, Peng Q. Expression of insulin-like growth factor binding protein 7 in gastric carcinoma and its clinical significance. Chinese J Digest 2012; 32: 559-560.
- Smith E, Ruszkiewicz AR, Jamieson GG, Drew PA. IGFBP7 is associated with poor prognosis in oesophageal adenocarcinoma and is regulated by promoter DNA methylation. Br J Cancer 2014; 110: 775-782.
- Yamashita S, Tsujino Y, Moriguchi K, Tatematsu M, Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci 2006; 97: 64-71.
- Rice JC, Ozcelik H, Maxeiner P, Andrulis I, Futscher BW. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis 2000; 21: 1761-1765.
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet 1999; 21: 163-167.
- Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr Biol 1997; 7: R305-307.
- Bird AP, Wolffe AP. Methylation-induced repression--belts, braces, and chromatin. Cell 1999; 99: 451-454.
- Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis 2000; 21: 461-467.
- Tycko B. Epigenetic gene silencing in cancer. J Clin Invest 2000; 105: 401-407.
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000; 16: 168-174.
- Ponder BA. Cancer genetics. Nature 2001; 411: 336-341.