Review Article - Journal of RNA and Genomics (2017) Volume 13, Issue 1
The Potential Link between Mobile DNA and the Invasiveness of the Species
Valeria Specchia1*#, Sally Janzen2, Gabriele Marini1 and Maurizio Pinna1*#
1Department of Biological and Environmental Sciences and Technologies, University of Salento, Italy
2School of GeoSciences, University of Edinburgh, EH9 3JN Edinburgh, UK
- *Corresponding Authors:
- Valeria Specchia
E-mail: valeria.specchia@unisalento.it
Tel: +39 0832 29 8673
Received: 29 May 2017; Revised: 20 June 2017; Accepted: 22 June 2017; Published: 28 June 2017
© Copyright The Author(s). First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
The environment and its stressors have an effect on the genome structure of species, affecting their adaptation and evolution. Environmental stresses significantly increase the genetic variability of a population and thus lay the basis for changes in fitness and unbalances. The movement of transposable elements represents a peculiar source of this variability in response to environmental changes and anthropogenic impacts. The relationship between stress and transposition has been exemplified in studies on different model organisms. Interestingly, mutational processes and genetic variation generated from transposable elements are emerging as potential agents of rapid adaptation of invasive species. In this research, we reviewed the role of transposable elements to reshape genomes with particular regards to RNAi processes in their control and potential relationship with the invasiveness of species.
Keywords
Transposable elements, Invasiveness, Genetic variation, Anthropogenic Impact, Epigenetic marks
Transposable Elements are Widespread Structural Components of the Genomes
Transposable elements are structural components and occupy large portions of all eukaryotic genomes. These DNA elements are repetitive sequences, potentially able to mobilize into the genome moving through RNA intermediates or using an autonomous mechanism of cut and paste (Bonchev and Parisod, 2013).
Transposable elements are peculiar in sequence and features across species. The majority of studies concern model organisms, such as Drosophila or mouse, and the human genome. The active human transposable elements are retrotransposons including Long Interspersed Nuclear Element 1 (LINE-1), Alu (a Short Interspersed Nuclear Element or SINE) and SVA. LINEs belong to a class of non-long terminal repeat retrotransposons (non-LTR) that move by a target-primed reverse transcription mechanism. SINEs are non-autonomous elements using other elements as a support for transposition (Prak and Kazazian, 2000).
The long-lasting study on the genome of the model organism Drosophila has highlighted the presence of a large number of families of transposons in its genome, including LTR and non- LTR retrotransposons and DNA transposons. The studies in this model have created the basis to understand the structure, the transposition mechanism, the control and the response to environmental changes of these elements (Brennecke et al, 2007; Palazzo et al, 2013; Piacentini et al, 2014).
More recently, many taxa of ecological significance begin to be investigated for the presence in the genome of transposable elements. Transposable elements, such as LINE-like, marinerlike and gypsy-like, but also SINE elements, have been reported for Mollusca species (Arkhipova and Meselson, 2000; Luchetti et al, 2016). The genome of the oyster Crassotrea virginica presents transposable elements belonging to the Miniature inverted–repeat transposable elements (MITEs)-like Pearl family (Gaffney et al, 2003). The bivalves Ruditapes decussatus and Ruditapes philippinarum represent the transposable element RUDI, a SINE element widely distributed in the genomes of a number of Mollusca species. In fact, due to their adaptation to different environmental conditions, the above mentioned species are farmed in aquiculture worldwide.
Transposable elements have also been characterized in different Crustaceans, including Daphnia, shedding light on the presence and structure of genomic mobile elements beyond the model organisms (Penton et al, 2002; Casse et al, 2006).
Transposable Elements are Shaping Components of the Genomes
The new insertions of transposable elements impact the global expression and functions of genes, resulting in effects at different levels of organization of living organisms (Biemont and Vieira, 2006; Feschotte and Pritham, 2007). At the cell level, transposable elements can insert into genes or their regulatory regions, potentially disrupting the gene function. Human diseases offer emblematic examples. In the brain of patients with schizophrenia new Line 1 insertions occur in or near genes related to synaptic functions and neuropsychiatric diseases (Bundo et al, 2014). The activity of transposons is deregulated in different cancer types and new insertions silence genes frequently mutated in cancer, including tumor suppressors (Lee et al, 2012). In Drosophila de novo gene mutations, produced by activated transposable elements, cause phenotypic variations (Specchia et al, 2010).
In addition to the inactivation of specific genes, transposons also act as extensive genome modifiers causing chromosome rearrangements through recombination events between multiple copies of their sequences (Lonning and Seedler, 2002). Overall the chromosome rearrangements globally create genetic variation within organisms, setting the basis for species evolution (Biemont and Vieira, 2006; Feschotte and Pritham, 2007).
Transposons may also act as gene regulatory elements providing promoters or causing a variation of the chromatin state (Sienski et al, 2012; Choung et al, 2017). The influence on the chromatin organization has large effects on gene expression because transposons act as nucleation points of condensed or heterochromatic states. Moreover these repetitive sequences contribute to create genomic domains characterized by particular chromatin states with specific epigenetic marks.
In the Drosophila model, the machinery of RNA interference (RNAi) is involved in the establishment of the chromatin state after transposon insertions. In particular, transposons nucleate histone marks, such as H3K9me3, with spread of these silencing marks in the flanking genomic regions. The resulting effect is a change in the expression of nearby genes (Wang and Elgin, 2011; Sienski et al, 2012; Le Thomas et al, 2013). Interestingly the epigenetic marks and related chromatin states are subject to variations in response to environmental stress (Seong et al, 2011). These changes can in turn cause the activation of transposons and novel events of structural modifications of the genome, establishing the basis for natural selection (Feil and Fraga, 2012).
Environmental Stressors Activate the Transposons Mobilization
Evidences of the activation of transposable elements by stress factors have been accumulated for several years in various organisms. By the late 1990, many studies demonstrated the activation of transposons in response to heat shock, irradiation and viral infections in the Drosophila model (Ratner et al, 1992, Anikeeva et al, 1994, Zabanov et al, 1995, Jouan-Dufournel et al, 1996; Vasil’eva et al, 1997). The effects of thermal stress on the activation of transposons have also been demonstrated in other organisms, such as the fungus Magnaporthe oryzae, a pathogen of the rice (Ikeda et al, 2001) and the ascomycete fungi Ophiostoma ulmi and Ophiostoma novo-ulmi, agents of the Dutch elm disease in humans (Bouvet et al, 2008).
The link between stress and transposable elements is exemplified by the extraordinary behavior of specific transposons in yeast (Dai et al, 2007). The Ty5 retrotransposon of yeast produces an integrate with a targeting domain (TD). The phosphorylated status of this domain allows the interaction with heterochromatin components and integrations in silent genes or gene-poor genomic regions. Interestingly, under stress conditions, such as starvation, the phosphorylation is reduced and the genome causing mutations is integrated. This fine regulation of Ty5 appears to form the mechanism to start the reshaping of the genome in critical conditions, allowing a potential adaptation (Dai et al, 2017). A fascinating example is also offered by the attitude of Line 1 elements to integrate in telomeres of mammal cells as a way to repair telomere-specific DNA damage (Morrish et al, 2007).
The molecular mechanisms underlying the activation of transpositions after stress are largely unknown, but the signals induced by stress could act on the processes devoted to controlling transposons in physiological conditions.
The Mechanisms Underlying the Transposable Elements Control Involve the RNA Interference
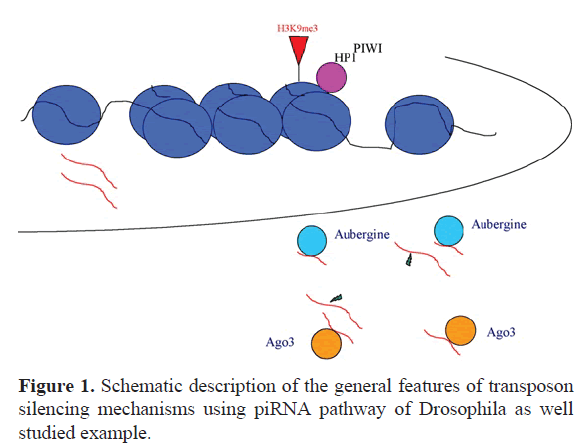
The silencing of transposable elements occurs at different levels of their life cycle, including transcriptional and post-transcriptional regulation. Different species take advantage of the DNA methylation as an epigenetic mechanism of transposable elements silencing. The methylation is a heritable mechanism to alter interactions between DNA and proteins, leading to changes in chromatin state and transcription rates. Conditions of hypomethylation lead to increased transposon activity in many taxa including mammals (Zamudio et al, 2015; Jones and Takai, 2001), plants (Law and Jacobnsen, 2010) and diatoms (Maumus et al, 2009). In the Drosophila model, the transcriptional silencing of transposons involves histone modifications. The methylation of histone H3 in K9 is the classical mark of transposon silencing mediated from the HP1, the protein involved in chromatin condensation and silencing. The RNAi pathway and the protein Piwi also act in the transcriptional silencing of these elements (Sienski et al, 2012).
The silencing also occurs at the post-transcriptional level and was widely studied, at first, in Drosophila. A specific RNAi pathway controls transposons after their transcription. It acts predominantly in gonads and uses a specific class of small non-coding RNAs, the piRNAs (Piwi interacting RNAs). The piRNA sequences correspond to a large amount of transposon’s classes of the Drosophila genome and their principal sources are genomic clusters containing sequence portions of transposons (Brennecke et al, 2007). The transcripts produced from clusters are transported in the cytoplasm to the specific sites of processing.
In the germ cells, the piRNAs are processed in a perinuclear region of the cytoplasm enriched with many proteins involved in transposon silencing (Zhang et al, 2012). In the somatic follicle cells of the ovary, the piRNAs are processed at the Yb bodies (Saito et al, 2010). Not only the sites, but also the key components differ between germ and somatic cells of the ovary.
In the germ cells, a “ping-pong” cycle performs piRNAs biogenesis and amplification. The Piwi clade proteins Aubergine (Aub) and Argonuate-3 (Ago3) are directly involved in this process and operate in a multi-protein perinuclear complex, the so called “nuage” (Brennecke et al, 2007). In the somatic follicle cells the piRNA-related factors are enriched in a spherical cytoplasmic region next to mitochondria, the Yb body, containing the tudor protein Yb (Szakmary et al, 2009) (Figure 1).
The piRNA pathway is conserved during evolution and acts in germinal cells of the mouse. In mammals, piRNAs are essential for DNA methylation of transposons (Aravin et al, 2008).
Environmental stress factors are able to induce changes in the methylation state of DNA and histones in different organisms (Seong et al, 2011; Feil and Fraga, 2012) and can lead to transposons activation.
The Potential Role of Transposons in the Genetic Basis of the Invasiveness
Invasive species are characterized by a rapid adaptation to new environments. This rapid and massive spread affects biodiversity and ecosystems functioning but also ecosystem services and human well-being, such as agriculture and fisheries (Mooney and Cleland, 2001; Pejchar and Mooney, 2009).
At the species level, common characteristics allow to the invasive species to colonize and spread rapidly in the new environment such as the high reproductive rate, long lived, high dispersion rate; in addition these species are generalists, pioneer species and devoid of natural enemies (Kolar and Lodge, 2001). At ecosystem level, invasive species can substitute the native species and, in the case of plants, determine changes in functional process (Fonnesu et al, 2004; Sangiorgio et al, 2004; Pinna et al, 2016), in biodiversity (Galuppo et al, 2007; Barbone et al, 2007) and in the ecological quality evaluation of aquatic ecosystems (Pinna et al, 2013; Orfanidis et al, 2007). Furthermore, substantial economic losses are reported due to invasions of agricultural pests, plant pathogens and competitors of fishes and aquatic species.
At the genetic level, the invasions could represent a consequence of a rapid genomic reshaping. Invasive populations are often based by a limited number of founding individuals and have a reduced genetic variability. Despite the low genetic variability, invasive species are successful leading to the concept of the genetic paradox of these species (Stapley et al, 2015).
From this point of view, transposable elements could represent the drivers of these genomic changes allowing adaptations (Casacuberta and Gonzalez, 2013). The movement of transposable elements, activated under conditions of environmental variability, creates rapid and extensive effects on gene activity and on the overall structure of genomes.
Their study is important to clarify the genetic and molecular basis of the invasiveness.
The alteration of their control in the germ cells ensures the transmission of the genome changes to the next generations. This fascinating hypothesis begins to find evidences, though not yet exhaustive. An example comes from aquatic invasive species. Invasive species of fishes belonging to the Cottus genus, hybrids between two lineages, present a large number of transposons copy variation. The changes in the transposon distribution may have created the genomic structural variation essential for the adaptation and invasion of new habitats (Dennenmoser et al, 2017).
Conclusions
Environmental stress conditions are the engine for the activation of transposable elements.
It is evident that the condition of stress unsettles the pathways of regulation and control of these elements. A paradigmatic example is the effect of stressors, such as heavy metals, dietary components and climate changes, on DNA methylation and epigenetic marks (Feil and Fraga, 2012). The relationship between stress conditions and other molecular transposon control mechanisms, such as the piRNA pathway, remain to be investigated.
The rearrangement of transposable elements could create the basis for adaptation and invasion in new and even extreme habitats. These studies will help to clarify which mechanisms are at the basis of the invasive capacity of species. The evolutionary conservation of the molecular pathways that control transposons indicates that could exist a common genetic mechanism of invasiveness. The resulting effects are species-specific in the relation to the peculiar reshaping of genomes performed by multiple actions of DNA mobile elements.
References
- Anikeeva NV, Zabanov SA, Vasil'eva LA, et al. 1994. Effect of heat shock on transpositions of mobile genetic element Dm412 in three isogenetic lines of Drosophila melanogaster. Genetika. 30, 212-217.
- Arkhipova A and Meselson M. 2000. Transposable elements in sexual and ancient asexual taxa. PNAS. 97, 14473–14477.
- Barbone E, Rosati I, Pinna M, et al. 2007. Taxonomic and dimensional structure of benthic macro invertebrate guilds in the Margherita di Savoia Salt Pans (Italy). Transit Water Bull. 1, 21-31.
- Biémont C and Vieira C. 2006. Genetics, Junk DNA as an evolutionary force. Nature. 443, 521-524.
- Bonchey G and Parisod C. 2013. Transposable elements and microevolutionary changes in natural populations. Mol Ecol Resour. 13, 765–775.
- Bouvet GF, Jacobi V, Plourde K, et al. 2007. Stress-Induced Mobility of OPHIO1 and OPHIO2, DNA transposons of the Dutch Elm Disease Fungi. Fungal Genet Biol. 45, 565-578
- Brennecke J, Aravin AA, Stark A, et al. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 128, 1089-1103.
- Bundo M, Toyoshima M, Okada Y, et al. 2014. Increased L1 Retrotransposition in the Neuronal Genome in Schizophrenia. Neuron. 81, 306-313.
- Casacuberta E and Gonzalez J. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22, 1503-1517.
- Casse N, Bui QT, Nicolas V, et al. 2006. Species sympatry and horizontal transfers of Mariner transposons in marine crustacean genomes. Mol Phylogenet Evol. 40, 609-619.
- Chuong EB, Elde NC and Feschotte C. 2017. Regulatory activities of transposable elements, from conflicts to benefits. Nat Rev Genet. 18, 71-86.
- Dai J, Xie W, Brady TL, et al. 2007. Phosphorylation Regulates Integration of the Yeast Ty5 Retrotransposon into Heterochromatin. Mol Cell. 27, 289-299.
- Dennenmoser S, Sedlazeck FJ, Iwaszkiewicz E, et al. 2017. Copy number increases of transposable elements and protein-coding genes in an invasive fish of hybrid origin. Mol Ecol. 1-13.
- Feschotte C and Pritham EJ. 2007. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu Rev Genet. 41, 331-368.
- Feil R and Fraga MF. 2012. Epigenetics and the environment, emerging patterns and implications. Nat Rev Genet. 13, 97-109.
- Fonnesu A, Pinna M and Basset A. 2004. Spatial and temporal variations of detritus breakdown rates in the river Flumendosa basin (Sardinia, Italy). Int Rev Hydrobiol. 89, 443-452.
- Gaffney PM, Pierce JC, Mackinley AG, et al. 2003. Pearl, a novel family of putative transposable Elements in Bivalve Mollusks. J Mol Evol. 56, 308-316.
- Galuppo N, Maci S, Pinna M et al. 2007. Habitat types and distribution of benthic macro invertebrates in a transitional water ecosystem, Alimini Grande (Puglia, Italy). Transit Water Bull. 1, 9-19.
- Ikeda K, Nakayashiki H, Takagi M, et al. 2001. Heat shock, copper sulfate and oxidative stress activate the retrotransposon MAGGY resident in the plant pathogenic fungus Magnaporthe grisea. Mol Genet Genomics. 266, 318-325.
- Kolar CS and Lodge DM. 2001. Progress in invasion biology, predicting invaders. Trends Ecol Evol. 16, 199-204.
- Jones PA and Takai D. 2001. The role of DNA methylation in mammalian epigenetics. Science. 293, 1068-1070.
- Jouan-Dufournel I, Cosset FL, Contamine D, et al. 1996. Transposable elements behavior following viral genomic stress in Drosophila Melanogaster Inbred Line. J Mol Evol. 43, 19-27.
- Law JA and Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 11, 204-220.
- Le Thomas A, Rogers AK, Webster A, et al. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 27, 390-399.
- Lee MM, Teuscher J, Miyasaka T, et al. 2012. Efficient hybrid solar cells based on meso-superstructured Organometal Halide Perovskites. Science. 338, 643-647.
- Lonning WE and Saedler H. 2002. Chromosome rearrangements and transposable elements. Ann Rev Genet. 36, 389-410.
- Luchetti A, Satovic E, Mantovani, B et al. 2016. RUDI, a short interspersed element of the V-SINE superfamily wiedespread in molluscan genomes. MOL GENET GENOMICS. 291, 1419-1429.
- Mateo L, Ullastres A and Gonzalez J. 2014. A Transposable element insertion confers xenobiotic resistance in Drosophila. PLoS Genet. 10, e1004560.
- Maumus F, Allen AE, Mhiri C, et al. 2009. Potential impact of stress activated retro transposons on genome evolution in a marine diatom. BMC Genomics. 10, 624.
- Mooney HA and Cleland EE. 2001. The evolutionary impact of invasive species. PNAS. 98(10), 5446–5451.
- Morrish TA, Garcia-Perez JL, Stamato TD, et al. 2007. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 446, 208-212.
- Orfanidis S, Papathanasiou V, Sabetta L, et al. 2007. Benthic macrophyte communities as bioindicators of transitional and coastal waters, relevant approaches and tools. Transit Water Bull. 1, 45-49.
- Palazzo P, Maggio P, Altavilla R, et al. 2013. Cerebral hemodynamics and systemic endothelial function are already impaired in well-controlled Type 2 diabetic patients, with short-term disease. PLoS ONE. 8, e83287.
- Penton EH, Sullender BW and Crease TJ. 2002. Pokey, a new DNA transposon in Daphnia (Cladocera, Crustacea). J Mol Evol. 55, 664-673.
- Pejchar L and Mooney HA. 2009. Invasive species, ecosystem services and human well-being. Trends Ecol Evol. 24, 497-504.
- Piacentini L, Fanti L, Specchia V, et al. 2014. Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma. 123, 345–354.
- Pinna M, Marini G, Rosati I, et al. 2013. The usefulness of large body-size macroinvertebrates in the rapid ecological assessment of Mediterranean lagoons. Ecol Indic. 29, 48-61.
- Pinna M, Marini G, Cristiano G, et al. 2016. Influence of aperiodic summer droughts on leaf litter breakdown and macroinvertebrate assemblages, testing the drying memory in a Central Apennines River (Aterno River, Italy). Hydrobiologia. 782, 111-126.
- Prak ETL and Kazazian HH. 2000. Mobile elements and the human genome. Nat Rev Genet. 1, 134-144.
- Ratner VA, Zabanov SA, Kolesnikova OV, et al. 1992. Induction of the mobile genetic element Dm-412 transpositions in the Drosophila genome by heat shock treatment. Proc. Natl. Acad. Sci. U.S.A. 89, 5650-5654.
- Saito K, Ishizu H, Komai M, et al. 2010. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24, 2493-2498.
- Sangiorgio F, Pinna M and Basset A. 2004. Inter- and intra-habitat variability of plant detritus decomposition in a transitional environment (Lake Alimini, Adriatic Sea). CHEM ECOL. 20, 353-366.
- Seong KH, Li D, Shimizu H, et al. 2011. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 145, 1049-61.
- Sienski G, Dönertas D and Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 151, 964-980.
- Specchia V, Piacentini L, Tritto P, et al. 2010. Hsp90 prevents phenotypic variation by suppressing the mutagenetic activity of trasposons. Nature. 463, 662-666.
- Stapley J, Sature AW and Tennis SR. 2015. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol. 24, 2241-2252.
- Szakmary A, Reedy M, Qi H, et al. 2009. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. JCB. 185, 613-627.
- Vasil'eva LA, Ratner VA and Bubenshchikova EV. 1997. Stress induction of retrotransposon transposition in Drosophila, reality of the phenomenon, characteristic features, possible role in rapid evolution. Genetika. 33,1083-1093.
- Wang SH and Elgin SCR. 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. PNAS. 108, 21164–21169.
- Zabanov SA, Vasil'eva LA and Ratner VA. 1995. Induction of transposition of MGE Dm412 using gamma-irradiation of an isogenic line of Drosophila melanogaster. Genetika. 31, 798-803.
- Zamudio N, Barau J, Teissandier A, et al. 2017. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256-1270.
- Zhang F, Wang J, Xu J, et al. 2012. UAP56 Couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 151, 871-884.