- Journal of RNA and Genomics (2013) Research Report
Sub-cellular temporal and spatial distribution of electrotransferred LNA/DNA oligomer
| Julie Orioα,β, Elisabeth Bellardα,β, Houda Baazizα,β, Chantal Pichonγ, Peter Mouritzen¥, Marie-Pierre Rolsα,β, Justin Teissiéα,β, Muriel Golzioα,β* and Sophie Chabotα,β* αCentre National de la Recherche Scientifique, Institut de Pharmacologie et de Biologie Structurale, BP64182, 205 route de Narbonne, F-31077 Toulouse, France βUniversité de Toulouse, UPS, IPBS, F-31077 Toulouse, France γCentre de Biophysique Moléculaire, CNRS UPR4301, rue Charles Sadron F-45071 Orléans, Cedex 02, Inserm and Université d’Orléans, France ¥Exiqon, Skelstedet 16, 2950 Vedbaek, Denmark *These authors contributed equally to this work. |
| *Correspondence to: Muriel Golzio, Email: muriel.golzio@ipbs.fr, or Sophie Chabot, Email: sophie.chabot@ipbs.fr, Tel: +33 561 175813/27, Fax: +33 561 175994 |
| Received 27 November 2012; Revised 04 March 2013; Accepted 15 March 2013; Published 15 March 2013 |
| © Copyright The Author(s): First Published by Library Publishing Media. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/ by-nc/2.5). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details. |
Abstract
Low biological activity and inefficient targeted delivery in vivo have hindered RNA interference (RNAi)-based therapy from realising its full clinical potential. To overcome these hurdles, progresses have been made to develop new technologies optimizing oligonucleotides chemistry on one hand and achieving its effective delivery on the other hand. In this report, we achieved, by using the electropulsation technique (EP), efficient cellular delivery of chemically-modified oligonucleotide: The locked nucleic acid (LNA)/DNA oligomer. We used single cell level confocal fluorescence microscopy to follow the spatial and temporal distribution of electrotransferred cyanine 5 (Cy5)-labeled LNA/DNA oligomer. We observed that EP allowed LNA/DNA oligomer cellular uptake providing the oligomer a rapid access to the cytoplasm of HeLa cells. Within a few minutes after electrotransfer, Cy5- LNA/DNA oligomers shuttle from cytoplasm to nucleus whereas in absence of pulses application, Cy5-LNA/ DNA oligomers were not detected. We then observed a redistribution of the Cy5 fluorescence that accumulated over time into cytoplasmic organelles. To go further and to identify these compartments, we used the HeLa GFPRab7 cell line to visualise late endosomes, and lysosomal or mitochondrial specific markers. Our results showed that the EP technique allowed direct entry into the cytoplasm of the Cy5-LNA/DNA oligomer bypassing the endocytosic pathway. However, in absence of pulses application, Cy5-LNA/DNA oligomer were able to enter cells through the endocytosic pathway. We demonstrated that EP is an efficient technique for LNA-based oligonucleotides delivery offering strong advantages by avoiding the endolysosomal compartmentalization, giving a rapid and free access to the cytoplasm and the nucleus where they can find their targets.
Keywords |
| Electropulsation, electroporation, RNAi, locked nucleic acid, fluorescence microscopy, cyanine 5 labeling |
Introduction |
| RNA interference (RNAi) is considered a major scientific breakthrough with potential therapeutic applications (Fire et al, 1998). Elbashir et al (2001) demonstrated the capacity of synthetic small interfering RNAs (siRNAs) to mediate RNAi in mammalian cells. Lagos-Quintana et al (2001) then demonstrated the identification in invertebrates and vertebrates of novel endogenous small non-coding RNA, termed microRNA (miRNA). Since then, miRNAs have been shown to regulate around 30% of the human genome and have been involved in a vast majority of physiological processes including development, cell differentiation, proliferation and apoptosis (Osman, 2012). Accordingly, altered miRNA expression is likely to contribute to human disease, including cancer (He et al, 2005). |
| AntagomiR is a small synthetic oligonucleotide that is perfectly complementary to its oncogenic miR target. This antisense strategy is currently used to silence endogenous oncogenic miRs. Overall, synthetic oligonucleotides interfering with the RNAi represented an important class of potential therapeutic pathway tools. |
| However, to date, the early promise of RNAi therapeutics has given way to some disappointment. In fact, firstly, classical oligonucleotides do not offer sufficient affinity and stability to be compatible with clinic and secondly, negatively-charged oligonucleotides do not permeate intact cell membranes, primarily because of the hydrophobic nature of the membrane lipid bilayer (Paganin-Gioanni et al , 2011). To improve oligonucleotides properties, chemically modified oligonucleotides have been developed including 2′-O-methyl, 2′-methoxyethyl, locked nucleic acids (LNA), and phosphorothiate linkages (Corey, 2007). Functional benefits of these backbone modifications are that it increases the thermodynamic properties of oligonucleotides. Thus, insertion of LNA bases into a DNA sequence (LNA/DNA oligomer) improves its base-pairing specificity and its resistance to nucleases and hence its efficacy (Kaur et al, 2007). Interestingly, we previously showed that electropulsation (EP) was an efficient technique for LNA-based oligonucleotide delivery (Chabot et al, 2012; Pelofy et al, 2012). EP is a physical method based on electric pulses application on cells inducing transient permeabilization of their membranes (Teissie et al, 2005). EP has been successfully used to increase cellular uptake of a wide variety of molecules such as plasmid DNA vector (Neumann et al, 1982; reviewed in Escoffre et al, 2009), cytotoxic drug (Mir et al, 1998; reviewed in Testori et al , 2012), siRNA (Paganin-Gioanni et al, 2008) and chemically-modified oligonucleotides (Chabot et al, 2011). In addition, we previously showed that electrotransferred LNA/DNA oligomer was efficiently silencing its target whereas in absence of pulses application no functional effect was observed (Chabot et al, 2012). However, the sub-cellular distribution of electrotransferred oligonucleotides remains unknown. |
| To be effective, therapeutic oligonucleotide has to find its target, meaning that sub-cellular localization of oligonucleotide is determinant for its biological effect. For instance, numerous reports have demonstrated that naked oligonucleotides are poorly internalized by cells and tend to localize in endosomes/lysosomes, where they are unavailable for their target (Braasch et al, 2002; Corsten et al, 2007; Alam et al, 2008; Juliano et al, 2008). In addition, a strong correlation between oligonucleotide nuclear localization and its specific efficiency has been reported (Marcusson et al, 1998; Zhang et al, 2011). Thus, knowledge about oligonucleotides dynamics and distribution in live cells is important for their transfer optimization. In the present paper, we studied the temporal and spatial sub-cellular localization of an oligomer containing LNA bases (LNA/DNA oligomer) during and after its electrotransfer. Using single cell confocal microscopy, we visualized the spatial and temporal bio-distribution of electrotransferred LNA/DNA oligomer which was labeled with the cyanine-5 (Cy5) fluorochrome. We observed that electrotransferred LNA/DNA oligomer had a direct access to the cytoplasm and gained the nucleus a few minutes after its electrotransfer where it stayed for a couple of hours. Our results showed that EP technique allowed efficient LNA/DNA oligomer delivery, avoiding endosomal and lysosomal compartment and offering strong advantages as compared to non-delivered oligonucleotides. In fact non-delivered oligonucleotides must exit from the endosome to reach its site of action in the cytosol or nucleus. Retention in endosomes must be avoided as oligonucleotides could activate immune response through Toll Like Receptors (TLRs). Moreover, electrotransferred LNA/DNA oligomer reaches its cytoplasmic and/or nuclear target rapidly (within a few minutes) which may limit its degradation by nucleases as compared to non-delivered oligonucleotides. By visualizing the temporal and spatial subcellular biodistribution of electrotransferred LNA/DNA oligomer, the present study highlights the interest of using EP for oligonucleotides delivery. Retention in endosomes must be avoided as oligonucleotides could activate immune response through Toll Like Receptors (TLRs). Moreover, electrotransferred LNA/DNA oligomer reaches its cytoplasmic and/or nuclear target rapidly (within a few minutes) which may limit its degradation by nucleases as compared to non-delivered oligonucleotides. By visualizing the temporal and spatial subcellular biodistribution of electrotransferred LNA/DNA oligomer, the present study highlights the interest of using EP for oligonucleotides delivery. Retention in endosomes must be avoided as oligonucleotides could activate immune response through Toll Like Receptors (TLRs). Moreover, electrotransferred LNA/DNA oligomer reaches its cytoplasmic and/or nuclear target rapidly (within a few minutes) which may limit its degradation by nucleases as compared to non-delivered oligonucleotides. By visualizing the temporal and spatial subcellular biodistribution of electrotransferred LNA/DNA oligomer, the present study highlights the interest of using EP for oligonucleotides delivery. electrotransferred LNA/DNA oligomer reaches its cytoplasmic and/or nuclear target rapidly (within a few minutes) which may limit its degradation by nucleases as compared to non-delivered oligonucleotides. By visualizing the temporal and spatial subcellular biodistribution of electrotransferred LNA/DNA oligomer, the present study highlights the interest of using EP for oligonucleotides delivery. electrotransferred LNA/DNA oligomer reaches its cytoplasmic and/or nuclear target rapidly (within a few minutes) which may limit its degradation by nucleases as compared to non-delivered oligonucleotides. By visualizing the temporal and spatial subcellular biodistribution of electrotransferred LNA/DNA oligomer, the present study highlights the interest of using EP for oligonucleotides delivery. |
Materials and Methods |
Cell lines and reagents |
| HeLa cell line was derived from human cervical adenocarcinoma. Cells were obtained from the America Type Culture Collection (ATCC) (ATCC Number CCL-2). HeLa cells were routinely maintained in Dulbecco's Modified Eagle Medium (Gibco-Invitrogen, Carlsbad, USA). HeLa GFP-Rab7 cell line was cultured in Eagle's Minimal Essential Medium with Eagle salt and non-essential amino acids (Eurobio, Les Ulis, France) added with 400μg/ml of G418 for keeping selection pressure. All media were supplemented with penicillin/streptomycin (100U/ml) (Eurobio) and 10% (w/v) heat inactivated fetal calf serum (Gibco-Invitrogen). Cells were maintained at 37°C in a humidified atmosphere with 5% (v/v) CO 2. Cells were mycoplasma free, as tested by enzymatic assay (MycoAlert, Lonza, Walkersville, MD, USA), and immunohistochemistry (DAPI staining). Singlestrand LNA/DNA oligomers labeled with cyanine 5 (Cy5) were supplied by Exiqon (Copenhagen, Denmark). The non-relevant LNA/DNA oligomer sequence was 5′-cagctcctcgcccttgctca-3′. Propidium iodide (PI) was obtained from Sigma Aldrich (Saint Louis, MO). |
Determination of cell permeability |
| For cells in suspension: cells (5 × 10 5) were harvested and resuspended in 100μl of pulsing buffer (10mM K2HPO4/ KH2PO4 buffer, 1mM MgCl2, 250mM sucrose, pH 7.4) in the presence of PI (100μM). Penetration of the PI was used to monitor cell permeability. The cell suspension was placed between 4mm-wide, stainless steel, flat, parallel electrodes (IGEA, Carpi, Italy) in a culture dish (Becton Dickinson, Rungis, France) and pulses lasting 5ms at a frequency of 1Hz were applied. EP was conducted using a S20u electropulsator (Betatech, l'Union, France), which delivered square-wave pulses with independently adjustable electric parameters (voltage, number of pulses, duration, and frequency). Pulse parameters were monitored in real time by the electropulsator LCD screen. |
| For cells in adherence: cells (15 × 10 4 ) were seeded on a 48 wells plate (Nalge Nunc International, Illkirch, France) overnight at 37°C in a humidified atmosphere with 5% (w/v) CO 2 . Medium was removed and 500μl of pulsing buffer containing PI (100μM) was added. Cells were pulsed using stainless steel, flat, parallel electrodes of 10mm-wide (IGEA). |
| After the electropulsation, PI fluorescence was analyzed by flow cytometry using a FACSCalibur (Becton Dickinson). A minimum of 104 events were acquired on the FL-2 channel and analyzed with CellQuest software (Becton Dickinson). |
Determination of cell viability |
| Cell viability was measured by quantifying cellular growth over 24hr (more than one generation) by crystal violet staining. Briefly, the cells were stained with 1ml crystal violet (0.1% (w/v) pulsing buffer) for 20min and then lysed with 500μl acetic acid (10% (w/v)) for 5min. Cell density was evaluated by 595nm OD measurements (Chabot et al, 2012). |
Confocal Fluorescence microscopy |
| For fluorescence microscopy observations, cells (8 × 10 4 ) were seeded on a glass coverslip chamber (Nalge Nunc International) 4hr before the experimentation in a humidified atmosphere at 37oC with 5% (w/v) CO 2. The electropulsation chamber was designed using two stainless steel parallel rods (1mm diameter, 10mm length and 4mm interelectrode distance) that were connected to the electropulsator. The chamber was set on the stage of an inverted confocal microscope (Zeiss LSM710, Carl Zeiss, MicroImaging GmbH, Göttingen, Germany) equipped with the 40x Zeiss objective (1.3 numerical aperture, oil immersion). Adherent cells were then pulsed in 500μl of pulsing buffer in the presence of 250nM (final) Cy5-labeled LNA/DNA oligomer or 100μM PI using the following electrical parameters: 300V/cm, 10 pulses of 5ms, 1Hz. Successive scans of less than 1sec were acquired (Zen 2009, Carl Zeiss) to observe the transfer of the molecule into cells before, during and after electric pulse delivery. Cy5 was visualized using a 633nm laser (emission filter: 640-710nm), GFP was visualized using a 488nm laser (emission filter: 492-570nm) and PI was observed using 488 laser (emission filter: 613-740nm). For long-term live cells imaging, cells were maintained under the microscope into a 37oC, 5% (w/v) CO2 incubation chambers. Acquisitions were performed each hour during 16hr. |
| Mitochondria and lysosome labeling and visualizations were performed 24hr after pulses application. For this purpose, cells were incubated for 30min at 37oC 5% (w/v) CO 2 with either 100nM of Mitotracker ® ​​Green FM (Invitrogen) or 50nM of Lysotracker ®Green DND-26 (Invitrogen) diluted in culture medium according to the manufacturer's protocol. Cells were washed once before confocal microscopy observation. 8-bit images were recorded with Olympus FV1000 software (Rungis, France) equipped with the 60x objective (1.4 numerical aperture, oil immersion). The laser scan was unidirectional and was perpendicular to the electric field direction to eliminate temporal delay during image acquisition. Cy5 was visualized using a 635nm laser (emission filter: 664nm), Mitotracker ® ​​Green FM and Lysotracker ® ​​Green DND-26 was visualized using a 488nm laser (emission filter: 520nm). |
Image analysis |
| LSM images were processed with ImageJ software (NIH, MD). For Cy5-LNA/DNA oligomer electrotransfer direct visualization, the fluorescence signal of the Cy5-labeled LNA/DNA oligomer in the cytoplasm of cells was analyzed by subtracting the first image (before EP) from each image of the scans series. This process eliminated the extracellular Cy5-labeled LNA/DNA oligomer signal (that was not affected by the electropulsation) and thus selectively discriminated the signal coming from inside the cells. |
Statistical analyzes |
| Quantitative data (represented as the means + sem) were analyzed with Prism 4 software (GraphPad, San-Diego, CA, USA). Two-way ANOVA were performed to statistically analyze effect of the field intensity on.cell permeability and viability. We report the actual P value for each test. |
Results |
Determination of efficient electrical parameters for in vitro cell electrotransfer |
| Adequate electrical parameters are dependent on the shape and size of the cells. Therefore, before performing an electrotransfer experiment, it is needed to determine the electrical condition leading to efficient permeabilization while preserving cell viability. Permeabilization of HeLa cells in suspension occurred only when field intensity was above a threshold value of 0.2kV/cm (Figure 1A). Field intensity significantly affected cell permeabilization and viability (p<0.0001 and p=0.0005, respectively). Permeabilization was enhanced whereas cell viability decreased as the field intensity increased. The optimal electric field intensity was determined as the highest percentage of viable permeabilized cells. It was set as 0.7kV/cm for cells in suspension (Figure 1A, dashed line). Plated cells permeabilization and viability were also dependent on field intensity (p < 0.0001). However, plated HeLa cells showed higher sensitivity for electric field. In this case, permeabilization started as the field intensity was up to 0.1kV/cm reaching a maximum value for a field intensity of 0.4kV/cm (Figure 1B). For plated cells, the optimal field intensity was obtained for a field intensity comprised between 0.3 and 0.4kV/cm (Figure 1B, dashed line). 1kV/cm reaching a maximum value for a field intensity of 0.4kV/cm (Figure 1B). For plated cells, the optimal field intensity was obtained for a field intensity comprised between 0.3 and 0.4kV/cm (Figure 1B, dashed line). 1kV/cm reaching a maximum value for a field intensity of 0.4kV/cm (Figure 1B). For plated cells, the optimal field intensity was obtained for a field intensity comprised between 0.3 and 0.4kV/cm (Figure 1B, dashed line). |
Visualization of Cy5-LNA/DNA oligomer electrotransfer at the single cell level |
| When plated HeLa GFP-Rab7 cells were pulsed using the optimum electrical parameters determined previously, cytoplasmic PI entrance was observed through sides of the cells facing the electrodes (Figure 2A). This result showed that the cell membrane was efficiently permeabilized at both sides. However, Cy5-LNA/DNA oligomer entry occurred solely at the side of the cell facing the negative electrode (cathode) (Figure 2B). This observation is consistent with previous experiments showing that oligonucleotides electrotransfer is dependent on the electrophoretic force (Golzio et al, 2002; Chabot et al 2012; Paganin-Gioanni et al, 2011). Cy5-LNA/DNA oligomer cellular uptake occurred as soon as the first pulse was applied (Figure 2B, 1p) and increased with the number of pulses (Figure 2B, 1p, 5p and 10p). Once the Cy5-LNA/DNA oligomer penetrated into the cells, it diffused into the cytoplasm (Figure 2B, 10p + 10sec) to finally get into the nucleus a few min after pulses application (Figure 2C). |
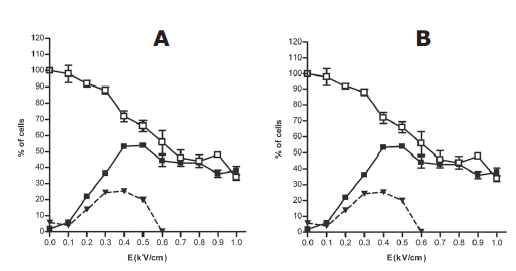 |
| Figure 1: Determination of optimum electric parameters on HeLa cells. Cells in suspension (A) or in adherence (B) were pulsed (10pulses of 5ms duration at 1Hz) with increasing field intensity (E in kV/cm). Cell viability was measured by crystal violet staining 24hr after pulse application (white square). Permeabilization was assayed by the penetration of PI into electrotransferred cells and analyzed by flow cytometry (black square). Values ​​are means ±SEM of three data sets. Percentage of viable permeabilized cells (dashed line) was calculated as follow: % (w/v) permeabilization + % (w/v) viability – 100. |
Cellular behavior of electrotransferred Cy5-LNA/DNA oligomer |
| We further followed over time the cellular localization of electrotransferred Cy5-LNA/DNA oligomer. 2Hr after the electrotransfer, the Cy5 fluorescence was observed mainly into the nucleus of pulsed cells but started also to be detected into the cytoplasm (Figure 3, 2hr). Then, the Cy5 fluorescence disappeared from the nucleus to concentrate over time into cytoplasmic organelles (Figure 3, 4hr-9hr). The confocal microscope settings were kept the same along the experiment to be able to compare Cy5 fluorescence intensity over time. It explains why we got saturation of Cy5 signal in areas where Cy5 concentration occurred. |
Sub-cellular distribution of Cy5-LNA/DNA oligomer |
| Twenty four hours after Cy5-LNA/DNA oligomer electrotransfer, cells were observed by confocal microscopy to identify the organelles in which the Cy5 fluorescence was found accumulated. To visualize the late endosomes, we used the HeLa GFP-Rab7 cell line. In fact, Rab 7 protein, which belongs to a superfamily of small molecular weight GTPases, is located specifically in the late endosomes. When cells were incubated in the presence of Cy5-LNA/DNA oligomer but without pulses application (non-pulsed cells; NP), we observed that the Cy5 fluorescence is partially colocalized with the GFP-Rab 7 fluorescence resulting in yellow pixels in the merge picture (Figure 4A, NP). On the contrary, no colocalization was observed when cells were pulsed in the presence of Cy5-LNA/DNA oligomer (Figure 4A,P). We then used the LysoTracker to label acidic organelles of HeLa cells. Once again, we observed that some of the Cy5 fluorescence colocalized with that of the LysoTracker in case of NP cells (Figure 4B, NP) whereas no colocalization was detected with pulsed cells (Figure 4B, P). Finally, we labeled mitochondria of HeLa cells using the MitoTracker. |
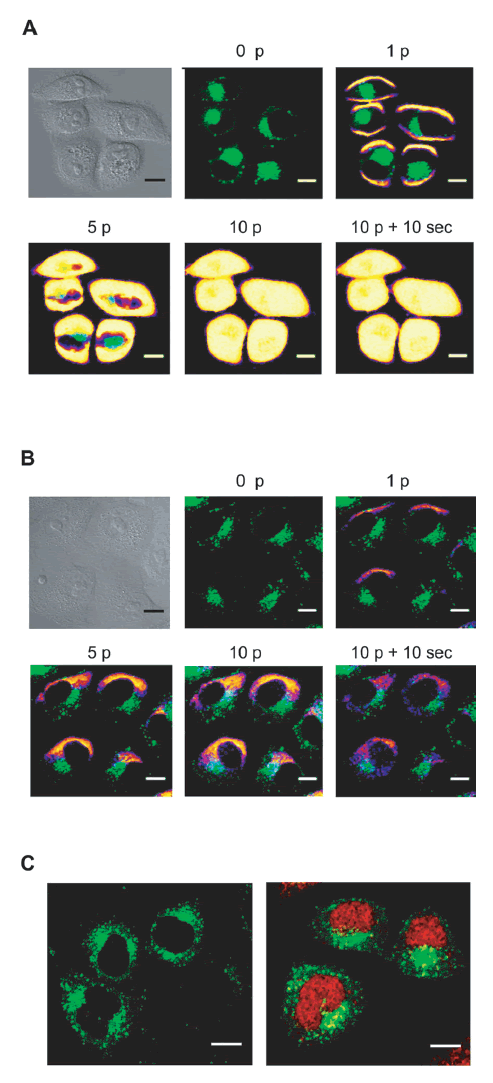 |
| Figure 2:Visualization of Cy5-LNA/DNA oligomer electrotransfer by direct confocal fluorescence microscopy. Plated Hela GFP-Rab7 cells were pulsed (0.3kV/cm, 10 pulses (p) of 5ms, 1Hz) on a glass coverslip chamber under a confocal microscope in the presence of PI (100μM) (A) or Cy5-LNA/DNA oligomer (250nM) (B). Scan series were carried out before (0p), during (1p, 5p and 10p) and 10sec after (10p + 10sec) pulses application. The anode was located on the bottom of the pictures. Scalebar = 10μm. Pictures of PI and Cy5 fluorescence are in pseudo-color, GFP fluorescence is in green and gray pictures are light transmission acquisition. C. Intracellular localization of Cy5-LNA/ DNA oligomers 2min after its application in non-pulsed cells (left picture) and in pulsed cells (right part). Scale bare = 10 μm. Cy5 and GFP fluorescence are in red and green respectively. |
| In NP cells, Cy5 fluorescence was strongly colocalized with that of MitoTracker, although not completely (Figure 4C, NP). In fact, some punctate Cy5 fluorescence was observed corresponding to lysosomal and/or endosomal compartment (Figure 4A and 4B, NP). When cells were pulsed in the presence of Cy5-LNA/DNA oligomer, Cy5 fluorescence exclusively colocalized with the MitoTracker signal (Figure 4C). |
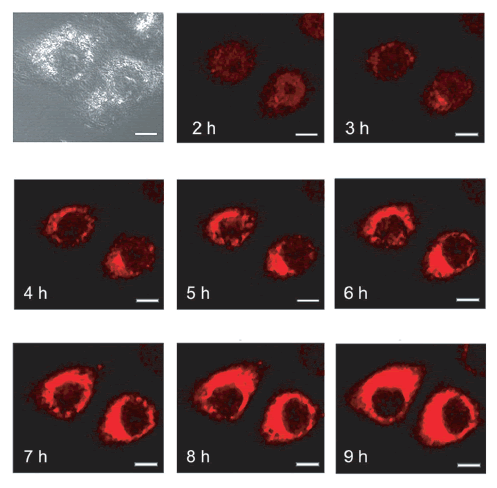 |
| Figure 3: Time course visualization of intracellular localization of electrotransferred Cy5-LNA/DNA oligomer. Plated HeLa cells were pulsed (0.3kV/cm, 10p of 5ms, 1Hz) on a glass coverslip chamber under a confocal microscope in the presence of 250nM of the Cy5-LNA/DNA oligomer and scan series were conducted every hour after the electrotransfer. Scale bar = 10μm. Pictures of Cy5 fluorescence are in red and gray picture is light transmission acquisition. |
Discussion |
| Chemically-modified oligonucleotides have been developed to overcome the limitation encountered with classical oligonucleotides, such as low affinity and low stability. Among this third generation of oligonucleotides, there is the LNA. Incorporation of LNA monomers in one strand of a nucleic acid duplex increases the melting temperature of the duplex with 2-8°C per LNA monomer. Thus, LNA vastly improves the thermal stability and specificity of duplexes formed with complementary DNA or RNA (Braasch and Corey, 2001). LNA oligonucleotides can be used in any application where high hybridization specificity is needed or when the target sequence is short. Thus, LNA-modified oligonucleotide appears well suited for RNAi therapeutic approaches (Elmen et al, 2005; Stenvang et al, 2008). However, just as all oligonucleotides, negatively-charged LNA-modified oligonucleotide cannot interact with the cell membrane that bears the same charge. LNA-modified oligonucleotide therefore cannot have free access to the cytoplasm. EP is an efficient delivery method to increase oligonucleotide cellular uptake but still it is necessary that access to its cellular target is effective. Interestingly, we previously demonstrated that electrotransferred LNA/DNA oligomer was functional, suggesting that it had access to its cellular target (Chabot et al, 2012). However, to our knowledge, LNA/DNA oligomer sub-cellular distribution after EP has never been published. In the present paper, we studied, by single cell confocal microscopy, |
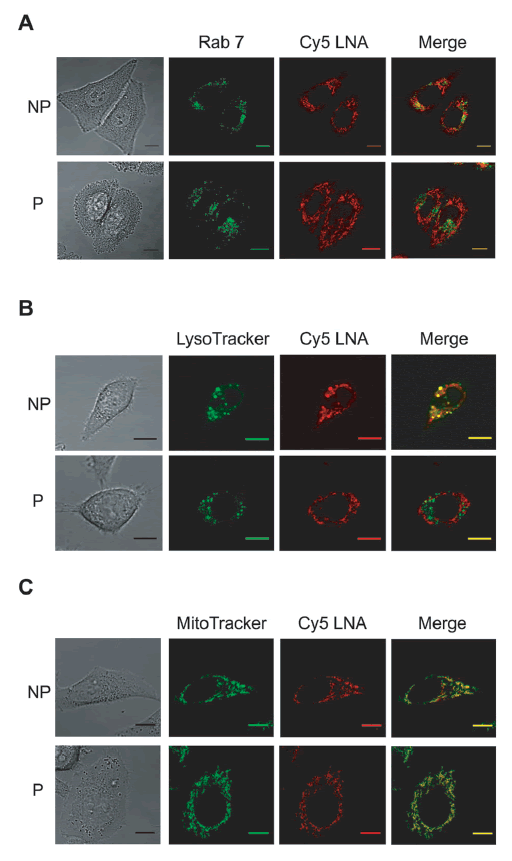 |
| Figure 4: Sub-cellular distribution of Cy5-LNA/DNA oligomer at 24hr. A. Plated HeLa GFP-Rab7 cells were pulsed (P) (0.3kV/ cm, 10 p of 5ms, 1Hz) or not (NP) in the presence of Cy5-LNA/ DNA oligomer (250nM). Confocal images were recorded 24hr after the treatment. Scale bar = 10μm. Representative pictures of GFP and Cy5 fluorescence are in green and red respectively and gray picture is light transmission acquisition. B and C. HeLa cells were treated as described above and labeled the following day either with Lysotracker ® ​​Green DND-26 (B) or Mitotracker ® ​​Green FM (C). Representative pictures of Lysotracker ® ​​Green DND-26 and Mitotracker ®Green FM fluorescence are in green, Cy5 fluorescence is in red and gray pictures are light transmission acquisition. Scale bar = 10μm. |
| Electropermeabilization occurs solely when the electric field intensity reaches a critical value that induces a sufficient transmembrane potential difference. This value is dependent on the size of the target; from 0.2kV/cm for large cells up to 1-2kV/cm in the case of bacteria (Teissie and Rols, 1993). Large cells are therefore more sensitive to lower field strengths than smaller ones. Therefore, electric field intensity must be adapted adjust to each cell line. We observed that HeLa cells permeabilization could be achieved by pulses application of field intensity above 0.2kV/cm for cells in suspension (Figure 1A) and 0.1kV/cm for adherent cells (Figure 1B). The optimum electric field intensity has been determined as 0.7kV/cm and 0. kV/cm for HeLa cells in suspension and in adherence respectively. This distinction is due to the difference in cell geometry of plated cells compared to cells in suspension and their increased sensitivity to the electric field (Valic et al, 2003). For the confocal microscopy experiments, HeLa cells were plated on a glass coverslip chamber so field intensity was maintained at 0.3kV/cm. |
| Using the electrical parameters determined above, we showed that HeLa cell membrane was permeabilized on the two opposite sides facing the electrodes (Figure 2A). Despite this double site of membrane permeabilization, entry of Cy5-LNA/DNA oligomer was only observed at the cathode side (Figure 2B). Cy5-LNA/DNA oligomer electrotransfer occurred as soon as the first pulse was applied increasing gradually with the number of pulses (Figure 2B). We thus confirmed that the electrotransferred Cy5-LNA/DNA oligomer entered directly, at the cathode side, into the cytoplasm through the permeabilized membrane driven by the electrophoretic forces. A few minutes after its electrotransfer, Cy5-LNA/DNA oligomer was located into the nucleus whereas no Cy5 fluorescence was detected in case of nonpulsed cells (Figure 2C). These results were in agreement with our previous observations (Chabot et al, 2012). |
| To go further, we then studied the spatial and temporal bio-distribution of the electrotransferred Cy5-LNA/DNA oligomer using living cells to prevent nonspecific artifact due to fixation protocol (Belitsky et al, 2002). In addition, working with living cells allowed us to follow over time the cellular distribution of electrotransferred Cy5-LNA/ DNA oligomer at a single cell level. Two hours after the electrotransfer, the Cy5-LNA/DNA oligomer was still present in the nucleus. Then, Cy5 fluorescence shuttled gradually to the cytoplasm to concentrate into undefined organelles (Figure 3). LNA/DNA oligomer efficacy has been correlated with its nuclear localization (Zhang et al, 2011) and more recently the nuclear retention phenomenon has been shown to give therapeutic advantage (Wheeler et al, 2012). Thus, by allowing LNA/DNA oligomer nuclear accumulation and its retention for at least a couple of hours (Figure 2C and 3), EP, in addition to be an efficient method of delivery, is likely to enhance oligonucleotides efficacy. |
| To identify the cytoplasmic organelles in which Cy5 fluorescence accumulates, we first used the HeLa GFP-Rab7 cell line that allows us to visualize late endosome (Decuzzi and Ferrari, 2008). In fact, standard lipofection systems enter cells through endocytosis; we wanted to know if this process was in some way involved with the technical EP. No Cy5 signal was found in this compartment in pulsed cells (Figure 4A,P). On the opposite, we detected some colocalization when Cy5-LNA/DNA oligomer was incubated with the HeLa GFP-Rab7 cells in absence of pulses application (Figure 4A, NP). To go deeper in our investigation, we either labeled HeLa cell lysosomes with the Lysotracker ®Green DND-26 (Figure 4B) or the mitochondria with the Mitotracker ®GreenFM (Figure 4C). For NP cells, we observed Cy5 fluorescence in lysosomal organelles whereas no Cy5 signal was detected in pulsed cells (Figure 4B). Finally, we observed that Cy5 fluorescence was strongly colocalized with that of MitoTracker, but not completely (Figure 4C, NP). In fact, some punctate Cy5 fluorescence was observed corresponding to lysosomal and/ or endosomal compartment (Figure 4A, B, NP). When cells were pulsed in the presence of Cy5-LNA/DNA oligomer, Cy5 fluorescence exclusively colocalized with the MitoTracker signal (Figure 4C). Unexpected observed mitochondrial colocalization is certainly due to non-specific accumulation of the Cy5 at this site as previously described (Lorenz et al, 2011). In fact, cationic dyes selectively accumulate in vitro and in vivo at mitochondria because of its high membrane electric potential (Rhee and Bao, 2010; Zhang et al, 2010). Therefore, Cy5-labeled oligonucleotides are not appropriate for long-time visualization study. The positive charge (e = +1) of the Cy5 should be counterbalance with the negative charges of the oligonucleotide, e = -20 in our case (-1 charge per nucleotide as insertion of LNA bases into DNA oligomer does not modify the negatively charged phosphodiester backbone). Thus, the overall net charge of the Cy5-LNA/DNA is negative, which should not cause its interaction with mitochondrial membrane. It is likely that the Cy5 release occurred in the nucleus and/or the lysosome, and that free Cy5 accumulates in mitochondria. Our results highlight the hurdle of using labeled-oligonucleotide, and especially cyanine, when long-time visualization is performed. |
| Taken together, our results showed that Cy5-LNA/DNA oligomer was able to enter cells by itself, probably through the endocytosic pathway as Cy5 fluorescence was detected in lysosomal and endosomal compartment (Figure 4A and 4B, NP). It is of note the Cy5 dye is not able to enter cells itself (Lorenz et al, 2011). On the contrary, the EP technique allowed direct entry of the Cy5-LNA/DNA oligomer into the cytoplasm bypassing the endocytosic pathway, which offers strong advantages. In fact, undelivered oligonucleotides firstly must undergo endosomal escape to reach their target before acidification in lysosomes, and secondly, may activate the endosomal Toll-like receptors, leading to unwanted immune activation (Heil et al, 2004). Thus, by bypassing the endocytotic pathway, EP improved the Cy5-LNA/DNA oligomer cellular uptake as well as its access to its cellular target. It has been published that gymnotic delivery was efficient to deliver chemically-modified oligonucleotides targeting mRNA (Stein et al, 2010). However, contrasting to gymnotic delivery, EP allowed rapid (within few minutes) and efficient intracellular delivery of small amount of long LNA-modified oligonucleotide and may be used to inhibit micro-RNA. In addition, EP offers the possibility to target different organs in vivo (Golzio et al, 2005; Golzio et al, 2007) whereas, intravenous injection (as used in unassisted delivery) mainly restricts delivery to the liver. It has been published that gymnotic delivery was efficient to deliver chemically-modified oligonucleotides targeting mRNA (Stein et al, 2010). However, contrasting to gymnotic delivery, EP allowed rapid (within few minutes) and efficient intracellular delivery of small amount of long LNA-modified oligonucleotide and may be used to inhibit micro-RNA. In addition, EP offers the possibility to target different organs in vivo (Golzio et al, 2005; Golzio et al, 2007) whereas, intravenous injection (as used in unassisted delivery) mainly restricts delivery to the liver. It has been published that gymnotic delivery was efficient to deliver chemically-modified oligonucleotides targeting mRNA (Stein et al, 2010). However, contrasting to gymnotic delivery, EP allowed rapid (within few minutes) and efficient intracellular delivery of small amount of long LNA-modified oligonucleotide and may be used to inhibit micro-RNA. In addition, EP offers the possibility to target different organs in vivo (Golzio et al, 2005; Golzio et al, 2007) whereas, intravenous injection (as used in unassisted delivery) mainly restricts delivery to the liver. EP allowed rapid (within few minutes) and efficient intracellular delivery of small amount of long LNA-modified oligonucleotide and may be used to inhibit micro-RNA. In addition, EP offers the possibility to target different organs in vivo (Golzio et al, 2005; Golzio et al, 2007) whereas, intravenous injection (as used in unassisted delivery) mainly restricts delivery to the liver. EP allowed rapid (within few minutes) and efficient intracellular delivery of small amount of long LNA-modified oligonucleotide and may be used to inhibit micro-RNA. In addition, EP offers the possibility to target different organs in vivo (Golzio et al, 2005; Golzio et al, 2007) whereas, intravenous injection (as used in unassisted delivery) mainly restricts delivery to the liver. |
Findings |
| Here we show that LNA-modified oligonucleotides electrotransfer may help to resolve RNAi problems relative to efficient target delivery and enhanced biological activity. This approach should lead to increased RNAi therapeutic efficacy and specificity. |
Acknowledgments |
| We thank Stéphanie Dauvillier (National Center for Scientific Research, Institute of Pharmacology and Structural Biology) for technical support. This work was supported by grants from the Seventh Framework European Program (FP7) OncomiR [grant number 201102], and was performed in collaboration with the “Toulouse Réseau Imagerie” core IPBS facility (Genotoul, Toulouse, France), which was supported by the Cancer Research Association (n°5585), Region Midi Pyrenees (CPER) and Grand Toulouse cluster. |
Competing Interests |
| None declared |
List of Abbreviations |
| RNAi; RNA interference |
| siRNA; small interfering RNAs |
| miRNA; microRNA |
| LNA; locked nucleic acids |
| Cy5; cyanine 5 |
| PE; electropulse |
References
|