Research Article - Biomedical Research (2017) Volume 28, Issue 20
Serum level of macrophage inflammatory protein 1 alpha predicts the response to pegylated interferon alfa-2a treatment in HBeAg positive chronic hepatitis B patients
Binfei Yang1, Ruifang Xu1, Caogeng Zhang1, Qin Zhang2 and Hui Wang3*
1Department of Infectious Diseases, Minhang Branch, Zhongshan Hospital, Fudan UniversityShanghai, China
2Department of Infectious Diseases, Tongren Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
3Department of Infectious Diseases, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- *Corresponding Author:
- Hui Wang
Department of Infectious Diseases
Ruijin Hospital
Shanghai Jiao Tong University School of Medicine
China
Accepted date: October 18, 2017
Abstract
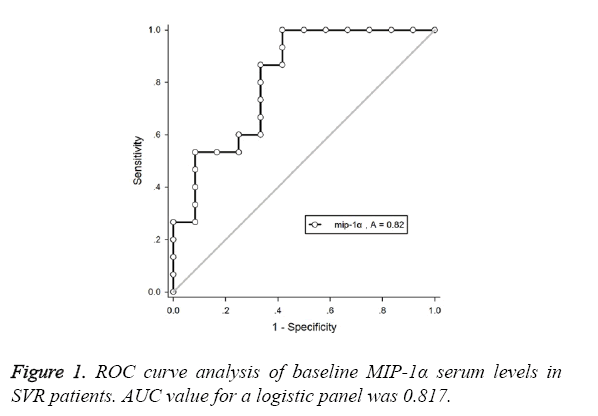
This study aimed to predict the response to pegylated interferon alfa-2a in HBeAg-positive Chronic Hepatitis B (CHB) by evaluating serum Macrophage Inflammatory Protein (MIP) 1α level. Total 31 patients with CHB were enrolled who were treated with PegIFNα-2a at a dose of 180 μg/w subcutaneously for 48 w. Serum MIP-1α level was measured at baseline and different period after PegIFNα-2a treatment. Baseline MIP-1α level of Sustained Virological Response (SVR) group (88.4 ± 21.3 pg/ml) was significantly lower compared to non-SVR group (163.4 ± 77.5 pg/ml). Female patients carrying genotype B, baseline HBV DNA ≤ 7l g IU/ml and baseline MIP-1α level ≤ 100 pg/ml at 12 w after treatment would be defined as easy responders. Those with serum MIP-1α level<110.979 pg/ml had the SVR positive predictive value of 98.065%, and with an area under the receiver operating characteristic curve of 0.817. In conclusion, low serum MIP-1α level is the predictor for the response of HBeAg-positive CHB patients to PegIFNα-2a treatment.
Keywords
PegIFNα-2a, Chronic hepatitis B, MIP-1α, Responder
Introduction
Chemokines promote hepatic inflammation and induce liver immunologic injury in Chronic Hepatitis B (CHB) infection through the recruitment of cytotoxic T Lymphocyte (CTL) to the infected liver [1]. Pegylated Interferon alfa (PegIFNα) based antiviral therapy is the first-line therapy for CHB, but has some disadvantages such as moderate antiviral effect, poor treatment tolerance, and frequent adverse effects. Predicting the response to PegIFNα-2a is the key to distinguish the patients who may benefit from PegIFNα-2a therapy from those who may not [2].
Among CXC chemokines, Macrophage Inflammatory Protein 1 alpha (MIP-1α) is a ligand for CXCR3 which is one of the receptors of Th1 associated chemokines [3,4]. MIP-1α could lead to viral clearance by regulating the differentiation of T helper cells, recruiting CD8+ T and B lymphocytes and inducing NK cells to produce IFN-γ and mediate inflammation [5]. MIP-1α is an inflammatory mediator that increases the secretion of other inflammatory mediators such as TNFα, IL-l and IL-6. MIP-1α activates the immune cells via the interaction with its corresponding receptor Chemokine Receptor 5 (CCR5), which plays a role in immune cell activation and migration in response to viral infections including hepatitis B. Therefore, assessment of CCR5 expression could help better understand the mechanisms responsible for the development of hepatitis B viral infection [6]. We speculated that chemokine MIP-1α would improve viral clearance by promoting the migration of T immune cells to CHB infected liver. In present study, we aimed to evaluate serum MIP-1α level in predicting the response to PegIFNα-2a in HBeAg-positive CHB patients.
Patients and Methods
Patients
This study was approved by institute ethics committee and informed consent was obtained from each participant. Total 31 HBeAg-positive chronic hepatitis B patients who received PegIFNα-2a at Infection Department of Shanghai Tongren Hospital and Ruijin Hospital from February 2007 to September 2012 were enrolled. The inclusion criteria were as follows: adults (22-48 y old), positive HBeAg statue for more than 6 months, HBV DNA levels higher than 105 IU/ml, and serum Alanine Amino-Transferase (ALT) level at 2-10 fold of the upper limit of normal level. The exclusion criteria were as follows: had epilepsy, heart disease, used drug, had hepatic disease such as HCV infection, liver cirrhosis, alcoholic liver disease, and autoimmune-mediated diseases, family psychiatric history, and were pregnant. The candidates with neutrophil count<1.0 × 109/L, platelet count<50 × 109/L and total bilrubin>51 μmol/L were also excluded.
Treatment and follow-up
The patients underwent monotherapy with PegIFNα-2a at a dose of 180 μg/w, administered subcutaneously for 48 w, and were followed up for 24 w.
HBV-M, HBV DNA, hepatorenal function and serum MIP-1α levels were evaluated at baseline, 12, 24, 36, 48 w of treatment and at 24-month follow-up. The virus gene types were identified at baseline.
According to Chronic hepatitis B Prevention Guide and the follow-up outcomes, patients were designated as Sustained Virological Response (SVR) or non-Sustained Virological Response (nSVR). SVR were defined as HBV DNA level lower than 500 IU/ml at 24 w of follow-up; nSVR were defined as HBV DNA level upper than 500 IU/ml at 24 w of follow-up.
Laboratory measurements
HBV-M level was determined by Abbott kit (USA) according to manufacturer’s instructions. Serum HBV DNA was quantified by PCR assay with a lower limit of detection of 500 IU/ml (Sai Le Qi GENE. Guangzhou, China), and serum ALT level was measured by a CX5 PRO Automatic chemistry system (Beckman, USA) with a upper limit of detection of 64 IU/L. MIP-1α was determined by MIP-1α ELLSA kit (Sen Xiong, Shanghai). HBV genotype was confirmed by genotyping kit.
Statistical analysis
Quantitative variables were expressed as mean and standard deviation. Categorical variables were presented as counts and percentages. The quantitative variables were analysed by t-test, and qualitative variables were analysed by Chi-square test or Fisher’s exact test. P-value<0.05 was considered significant.
Results
Patient characteristics
A total of 31 patients including 17 SVR and 14 nSVR patients were enrolled. The average age of SVR group and nSVR group was 30.5 ± 3.9 y and 36.1 ± 5.3 y, respectively. The average age of SVR group was significant lower than nSVR group (p=0.003). The male ratio of SVR group (6/17, 35.29%) was lower than nSVR group (11/14, 78.57%). Among the 31 patients, there were 11 (64.7%) cases of genotype B in SVR group and 3 (21.4%) cases of genotype B in nSVR group, showing significant difference between two groups. The level of HBV DNA was significantly lower in SVR patients with HBV DNA level of 6.48 ± 0.79 IU/ml than in nSVR patients with HBV DNA level of 7.22 ± 0.63 IU/ml (p=0.014). The level of baseline MIP-1α was significantly lower in SVR patients (88.4 ± 21.3 pg/ml) than in nSVR patients (163.4 ± 77.5 pg/ml) (p=0.006). These results showed that the younger female who had the lower levels of HBV DNA and MIP-1α and carried genotype B with shorter time of course of disease would be defined as SVR.
Single-factor and multi-factor analysis of the factors predicting response to PegIFNα-2a treatment
To analyse the efficacy of PegIFNα-2a treatment, we performed single-factor analysis and found that the females with genotype B had a HBV DNA level ≤ 7l IU/ml. MIP-1α level at 12 w post-treatment ≤ 100 pg/ml was used to define the responders (Table 1). However, those parameters at 24 w posttreatment determined by multi-factor analysis could not be the predicting factors (p>0.05).
| Single factor analysis | Multi factor analysis | ||||||
|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95% CI | P | ||
| Predictive factors | |||||||
| Age (y) | >35 vs.≤35 | 7.5 | 1.228-45.806 | 0.0291 | NS | ||
| Gender | Male vs. female | 6.722 | 1.332-33.913 | 0.021 | 13.569 | 0.814-226.315 | 0.0693 |
| Course of disease (month) | >36 vs.≤36 | 2.571 | 0.598-11.058 | 0.2045 | NA | ||
| HBV Genotype | B vs. C | 0.149 | 0.029-0.750 | 0.021 | 0.076 | 0.004-1.531 | 0.0926 |
| ALT (U/L) | >200 vs.<200 | 1.375 | 0.322-5.877 | 0.6674 | NA | ||
| HBV-DNA (lgIU/ml) | >7vs.≤7 | 6 | 1.261-28.547 | 0.0244 | 2.574 | 0.199-33.359 | 0.4695 |
| HBsAg (lgIU/ml) | >4000 vs.≤4000 | 2 | 0.396-10.089 | 0.4013 | NA | ||
| HBeAg (lgS/CO) | >1000 vs.≤1000 | 1.806 | 0.377-8.640 | 0.4595 | NA | ||
| MIP-1α (pg/ml) 0w | >100 vs.≤100 | 8 | 1.399-45.756 | 0.0194 | 6.848 | 0.401-116.945 | 0.1839 |
| MIP-1α (pg/ml) 12w | >100 vs.≤100 | 11 | 1.103-109.674 | 0.041 | NS | ||
| OR: Odds Ratio; NA: Not Adopted; NS: Not Significant. | |||||||
Table 1: Single-factor and multi-factor analysis of predicting response factors at 24 w post-treatment.
MIP-1α serum level could be a predictor of SVR HBeAg-positive CHB patients to PegIFNα-2a treatment
As shown in Table 2 and Figure 1, SVR patients with baseline MIP-1α level<110.979 pg/ml had a Positive Predictive Value (PPV) of 96.065%, and the corresponding area under the ROC curve was 0.817.
| Predictors | AUC | Youden index | PPV | NPV | Cut-off |
|---|---|---|---|---|---|
| Baseline MIP-1α (pg/ml) | 0.817 | 0.6359 | 0.96065 | 0.4702 | 110.979 |
| W 12 MIP-1α (pg/ml) | 0.804 | 0.55 | 0.85715 | 0.66665 | 132.996 |
Table 2: MIP-1α predicts SVR HBeAg-positive CHB patients with PegIFNα-2a treatment.
ALT recover, HBV DNA negative conversion, and HBeAg seroconversion in the treatment and follow-up between the responders and non-responders
According to the response to ALT recover, HBV DNA negative conversion and HBeAg seroconversion, among 31 CHB patients 6 (19.35%) were responders and 22 (70.97%) were non-responders. The patients with lower level of baseline HBeAg could be easily defined as responders. ALT recover, HBV DNA negative conversion and HBeAg seroconversion were not significantly correlated with the age, gender, course of disease, HBV genotype, baseline HBV DNA, ALT, and MIP-1α level of CHB positive patients (Table 3).
| SVR group | nSVR group | χ2 (Z) | P | |
|---|---|---|---|---|
| Age (y) | 30.22±4.44 | 34.23±5.31 | -1.8112 | 0.0701 |
| Male (n, %) | 3(17.6) | 14(82.4) | 0.9742 | 0.2329 |
| Course of disease (month) | 45.56±39.98 | 65.64±42.05 | -1.4223 | 0.1549 |
| Genotype | 0.3235 | 0.6424 | ||
| B n (%) | 7(50.0) | 7(50.0) | ||
| C n (%) | 2(11.8) | 15(88.2) | ||
| ALT (U/L) | 217.44±99.17 | 197.05±72.59 | 0.3267 | 0.7439 |
| HBV-DNA (lgIU/ml) | 6.42±0.76 | 6.97±0.78 | -1.7214 | 0.0852 |
| HBsAg (lgIU/ml) | 3.79±0.38 | 3.77±0.24 | 0.1089 | 0.9133 |
| HBeAg (lgIU/ml) | 2.32±0.58 | 2.80±0.41 | -2.2646 | 0.0235 |
| MIP-1α (pg/ml) | 80.12±17.59 | 132.48±72.22 | -1.8535 | 0.0638 |
Table 3: Comparison of SVR group and nSVR group at 24 w posttreatment.
Discussion
Identification of the predictors for predicting virological response to the treatment is important to guide the options of antiviral drug for the treatment of viral infection. In this study, for the first time we reported that the chemokine MIP-1α serum level could predict the response to Pegylated IFNα-2a therapy in patients with CHB.
Heydtmann et al. reported that HCV-induced secretion of CCR5 attracted immature plasmacytoid Dendritic Cells (pDCs) to the liver to influence HCV pathogenesis. HCV infected liver cells then trigger the secretion of chemokines, including MIP-1α and MIP-1β, which amplify the recruitment of CD4 T cells [7]. In our study, the percentage of responders to PegIFNα-2a in CHB patients with lower baseline MIP-1α level was higher. A possible explanation could be that decreased number of CHB-positive liver cells after PegIFNα-2a treatment would induce a low level MIP-1α production. The responders had a significant increase of MIP-1α level from baseline at the 12 w post-treatment compared to non-responders. These findings are consistent with previous study that the responders of CHC patients had higher MIP-1α level after 24 h of IFNα and ribavirin combined treatment [8]. However, it is still unclear how increase in MIP-1α level results in decreased response to PegIFNα-2a treatment in HBeAg-positive CHB patients. Further mechanistic studies are needed. In addition, the changes in CCR5 expression pattern in CHB patients during PegIFNα-2a treatment should be characterized to reveal the mechanism by which MIP-1α plays a role in immunological response to HBV infection.
Due to the limitation of small sample size of this study, further large-scale studies are needed to confirm that serum MIP-1α level could predict the response to PegIFNα-2a treatment in CHB patients.
In summary, our study suggests that the increase of MIP-1α level of CHB patients indicates the decreased percentage of long-term response for the management of CHB. Baseline MIP-1α serum level could be used as a predictor of sustained response to PegIFNα-2a treatment in HBeAg-positive CHB patients.
Conflict of Interest
No
References
- Wang J, Wang PP, Xiang GJ. Relationship between the expression of IP-10 and IP-10 mRNA in peripheral blood and HBV DNA level in patients with cirrhosis. HepatobiliaryPancreat Dis Int 2010; 9: 280-286.
- Brunetto MR, Lok AS. New approaches to optimize treatment responses in chronic hepatitis B. AntivirTher 2010; 15: 61-68.
- Liaskou E, Wilson DV, Oo YH. Innate immune cells in liver inflammation. Mediators Inflamm 2012; 2012: 949157.
- Harvey CE, Post JJ, Palladinetti P. Expression of the chemokine IP-10 (CXCL10) by hepatocytes in chronic hepatitis C virus infection correlates with histological severity and lobular inflammation. J Leukocyte Biol 2003; 74: 360-369.
- Zeremski M, Petrovic LM, Talal AH. The role of chemokines as in ammatory mediators in chronic hepatitis C virus infection. J viral Hepat 2007; 14: 675-687.
- Sanchooli J, Sanadgol N, KazemiArababadi M, Kennedy D. CCR5 plays important roles in hepatitis B infection. Viral Immunol 2014; 27: 2-6.
- Heydtmann M, Adams DH. Chemokines in the immunopathogenesis of hepatitis C infection. Hepatology 2009; 49: 676-688.
- Florholmen J, Kristiansen MG, Steigen SE. A rapid chemokine response of macrophage inflammatory protein (MIP)-1alpha, MIP-1beta and the regulated on activation, normal T expressed and secreted chemokine is associated with a sustained virological response in the treatment of chronic hepatitis C. ClinMicrobiol Infect 2011; 17: 204-209.