Research Article - Microbiology: Current Research (2018) Volume 2, Issue 3
Screening, isolation, biochemical and plant growth promoting characterization of endophytic bacteria.
Abid Ullah1, Hafsa Mushtaq2, Usman Ali1, Hakim1, Ehsan Ali3, Samavia Mubeen2
1Department of Plant Science and Technology, Huazhong Agricultural University, Wuhan, Hubei, P. R. China
2Department of Plant Sciences, Faculty of Biological Sciences, Quaid-i-Azam University, Islamabad, Pakistan
3Hubei Insect Resources Utilization and Sustainable Pest Management Key Laboratory, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, P. R. China
- *Corresponding Author:
- Hassan Javed Chaudhary
Department of Plant Sciences
Faculty of Biological Sciences
Quaid-i-Azam University
Islamabad, Pakistan
E-mail: abidqau.101@gmail.com
Accepted date: February 23, 2018
Abstract
Microbially unexplored medicinal plants can have a genetically diverse microbial population with multi-functional plant growth promoting traits. Altogether, 75 endophytic bacterial strains were isolated from Withania coagulans Dunal and Olea ferruginea Royal, medicinal plants of Karak, Pakistan. Isolated endophytic bacterial strains were grouped into 8 clusters while, 4 remained ungroup on the basis of the SDS-PAGE profile of whole cell protein. Clusters representative bacterial strains were evaluated for different activities, among them 4 strains were selected for 16s rDNA gene sequencing due to their prominent results. The 16s rDNA gene sequencing homology of the representative strain WC2, WC58, WC138 and OFR115 were assigned to Enterobacter cloacae, Enterobacter dissolvens, Enterobacter hormaechei and Cronobacter Sakazakii respectively. Phylogenetic tree indicated evolutionary relationship of these bacteria to their closely related species. All of the representative bacterial isolates were screened for biochemical characteristics, and various plant growth promoting activities like phosphate solubilization, ammonia production, and indole-3-acetic acid (IAA). Indirect plant growth promoting traits, such as hydrogen cyanide production, protease production, salt tolerant capability and antagonistic activities against plant pathogenic fungi under in vitro conditions were also evaluated. These isolates were also characterized biochemically. Furthermore, the isolation of endophytic bacteria from Withania coagulans and Olea ferruginea is novel in association to these plants. Owing to these characters, the representative isolated bacteria are suggested to be used in bio-fertilizer and bio-pesticides.
Keywords
Biocontrol agents, Endophytic bacteria, Plant-growth-promoting traits, Symbiosis.
Introduction
Endophytic bacteria by definition are those bacteria that colonize the internal tissue of the plants either symbiotically or in a mutualistic relationship [1]. Likewise, soil carries a rich pool of microbial inoculum for rhizosphere endophytes that readily colonize and exclusively invade host tissues. The microbial community isolated from rhizosphere is of special importance to the plants [1]. Therefore, endophytes that infect plants from the soil must be competent root colonizers. The variation in the endophytic communities can be attributed to bacterial species, plant genotype, plant developmental stage, colonizing tissue type, soil type and environmental conditions [2]. Monocotyledonous as well as dicotyledonous plants and their compartments (i.e. roots, stem, leaves, seeds) have been explored for endophytic bacteria [3]. Different endophytes have been identified from different plant tissues. The biodiversity of endophytes can be judged by the potential of microbes to colonize the tissues and persist in the plant [4]. Molecular diversity of endophytic bacteria in a single plant is not limited to a single species, but comprises a number of different genera and species. It may be varied even in different tissues of a single plant [5]. In general, endophytic bacterial populations in plant tissues have been reported between 102 to 104 viable bacteria per gram [2].
Bacteria that can fix nitrogen i.e. convert stable atmospheric nitrogen gas into a biologically useful form are known as diazotrophs. These organisms reduce di-nitrogen to ammonia with the help of enzyme, nitrogenase [6]. Diazotrophic bacteria can positively influence plants by improving growth and root development, which increases plant tolerance to various environmental stresses [7]. All plants naturally harbour a diverse range of endophytic bacteria which can affect positively host growth along with soil quality. Variations in plant growth frequently reflect alterations in phytohormones levels induced by endophytes [1]. Endophytes are well known for their potential to improve plant growth by direct and indirect mechanisms. Direct mechanisms involve the microbial synthesis of phytohormones for example, production of Indole-3-acetic acid (IAA), ethylene like, cytokinins like and gibberellins like. In addition, these endophytic bacteria also have the ability of nitrogen fixation [8]. Moreover, the endophytic bacteria have the potential to produce 1-aminocyclopropane-1-carboxylate (ACC) deaminase, which lowers plant ethylene levels because ACC is a precursor for ethylene production [9,10]. On the other hand, indirect mechanisms include suppression of pathogen infection via antifungal or antibacterial agents. The other indirect mechanisms include assisting plants in acquiring nutrients via phosphate solubilisation, nitrogen fixation and siderophores production. Besides these mechanisms, plant-associated microorganisms improve nutrient acquisition by supplying minerals and other micro/macro nutrients from the soil [11,12]. Therefore, isolation and characterization of endophytic bacteria with various properties from unexplored hosts will have much applications to improve plant growth promotion [13,14].
In the present study, Withania coagulans and Olea ferruginea were selected on the basis of their wide medicinal importance. The aim of the present study was to isolate, find genetic diversity and characterize plant growth promoting (PGP) potential of endophytic bacteria from Withania coagulans and Olea ferruginea.
Materials and Methods
Isolation of bacterial endophytes
The plants were collected in August of 2013 from Sabir abad, district Karak, Khyber Pakhtun Khwa, Pakistan (33.14618 latitude, 71.18926 longitude) as the source material for bacterial endophytes isolation. Surface disinfection was conducted by using serial washing with 70% ethanol for 5 min, 0.1% HgCl2 for 1-2 min and then double distilled water, 5-6 times for 2-5 min. A 100 μl sample of the water from the third rinse was plated on 869 medium [15] to verify the efficiency of sterilization. Three replicates of each sterilized plant part samples were transferred into test tubes containing 5 ml semisolid medium. These samples were incubated for 7 days at 28°C and observed for the growth of endophytic bacteria. Morphologically different colonies were selected and purified on LB agar plates.
SDS-PAGE of whole-cell crude protein
The methods of cell preparation, protein extraction, data analysis and gel electrophoresis was followed as described by [16].
Biochemical characterization of endophytic bacteria
Bacterial cultures were screened for physiological and biochemical characters through molecular identification kits, QTS 24 miniaturized identification system (DESTO Laboratories Karachi, Pakistan). Bacteria were cultured for 24 h in nutrient broth medium at 35 ± 2°C to inoculate QTS kits.
Phosphate solubilization by Endophytes
Bacterial isolates were screened for their potential to solubilize insoluble calcium phosphate on Pikovskaya agar medium as described by Pikovskaya, [17]. Bacterial colony was placed on the center of Pikovskaya agar medium plates with the help of sterile loop and incubated at 30°C for 7 days. The phosphate solubilizing efficiency was measured on the basis of halo zones around the colonies by following the formulae of [18,19] and was measured by:
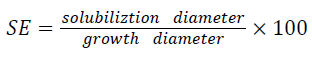
Quantitative estimation of Indole-3-Acetic Acid (IAA)
Production of Indole-3-acetic acid (IAA) was estimated for all bacterial isolates by inoculating 500 μl of 24 h old bacterial suspension in 50 ml of broth containing 0.1% DL-tryptophan and kept in a shaker incubator at 30 ± 2°C for 5 days at 180 RPM in the dark. IAA concentration in the culture supernatant was estimated by mixing 4 ml of salkowski reagent in 1 ml of supernatant and absorbance of the resultant pink color was read after 30 min at 535 nm in UV/Visible Spectrophotometer. The IAA production was measured from the regression equation of a standard curve and the result was expressed as μg ml-1 [20].
Ammonia production
Bacterial endophytes were tested for the production of ammonia in peptone water. Freshly grown cultures were inoculated in 10 ml peptone broth in each tube and incubated at 28 ± 2°C for 48-72 h. Nessler’s reagent was added to each tube after incubation. The development of faint yellow to dark brown color was a positive test for ammonia production [21].
Screening of isolates for HCN
Qualitative determination of hydrogen cyanide (HCN) for all bacterial isolates were carried out by adapting the method of Lorck with few modifications [22]. Bacterial isolates were streaked on nutrient agar medium supplemented with 4.4 gl-1 of glycine. The production of cyanide was detected by placing Whatman filter paper No.1 soaked in 0.5% picric acid under side of the petri dish lids. Development of brown to red color after incubation of four days indicated HCN production.
Antifungal activity
Overnight grown bacterial culture was streaked on the four ends of the petri plates on SDA (Sabouraud dextrose agar) medium and spores of fungal species Macrophomia phaseolina (ATCC62589) were spot inoculated at the center of plates and incubated at 28 ± 2°C for 5-8 days. Antifungal activity was noted as the prohibited growth of mycelium in the direction of bacterial colony [23]. Percent of radial growth inhibition was recorded by the following formula:
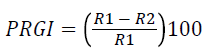
PRGI: Percent of radial growth inhibition, R1: Radius of fungus, R2: Radius of bacterial colony
Assessment of bacterial isolates for enzymatic activity
The 24 h old bacterial colony was mixed with a drop of 3% hydrogen peroxide on a glass slide by using a sterile toothpick or a loop. The formation of bubbles considered positive for catalase activity. To screen amylase activity, 24 h old bacterial cultures were spot inoculated on starch agar plates and incubated at 28 ± 2˚C for 48 h. After proper incubation, plates were flooded with gram iodine solution. Appearance of the clear yellow zone around the bacterial growth indicated starch hydrolysis while blue zone surrounding the bacterial growth, indicated that starch is present and has not been hydrolyzed [24]. To measure protease enzyme, fresh cultures of bacterial isolates were placed on the center of the SMA medium (15 g skim milk, 0.5 g yeast extract, 9.3 g agar and 1 L distilled water) and incubated for 48 h. Colorless halo zones around bacterial colonies were indicative of protease production.
Determination of growth at different salt concentration
Bacterial isolates were checked for salt tolerance at various concentrations. Fresh cultures were streaked on nutrient agar medium containing 0%, 7.5%, 10% and 15% NaCl. After 24 h of incubation at 28 ± 2˚C results were noted as positive or negative growth.
16s rDNA gene sequencing of selected isolates
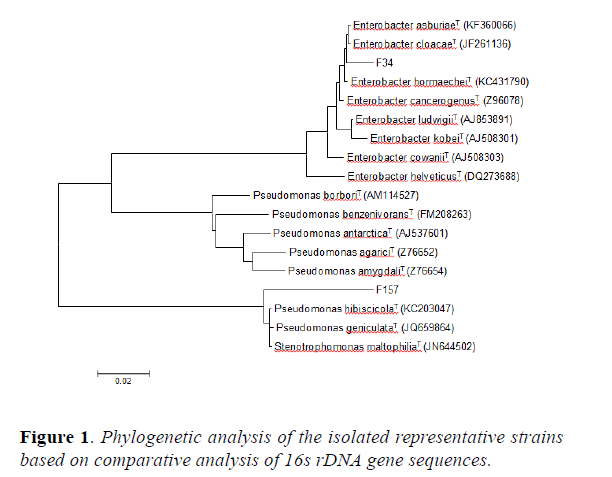
Fragments of the 16s rDNA gene were amplified from genomic DNA of the isolates. The amplification and PCR-based direct sequencing procedure were followed by [5]. The acquired sequences, together with some related sequences, were aligned with FASTA3 programe [25]. Ambiguous bases were excluded from the calculation of similarity. The tree topology was inferred by the neighbor joining method and the phylogenetic tree was visualized and bootstrapped 1,000 times of resampling by using the TRECON software package [26].
Results
Isolation and SDS-PAGE of whole cell proteins
Seventy five bacterial strains were isolated from root, stem and leaves of W. coagulans and O. ferruginea. Protein profile was studied by SDS-PAGE analysis for all isolated bacteria of both plants. The strains that showed similar protein profiles were grouped in the form of clusters which based on visual comparison of band pattern. Each group has a similar band pattern, but differ from other group. Out of 75 isolated bacterial strains, 71 were grouped into 8 clusters (I-VIII) while 4 remained ungroup since their band pattern did not match to any group (Appendices S1).
Biochemical characterization of bacterial endophytes
The biochemical characteristics of isolated strains were tested by microbial identification kits QTS-24. The test showed the utilization of different nitrogen and carbon sources by the tested strains. The utilized carbon/nitrogen sources by these representative strains shown in Appendices 2 and described in Table 1.
| Isolated representative strains | F34 | F157 |
|---|---|---|
| Phosphate solubilization | 1.8 | - |
| Ammonia production | + | + |
| HCN production | + | - |
| IAA production (µg) | 12.3 | 10.3 |
| MIC for Pb (ppm) | 1150 | NA |
| MIC for Cd (ppm) | 900 | NA |
| MIC for Hg (ppm) | NA | 100 |
Table 1: PGP and heavy metals characterization of isolated representative strains.
In vitro screening of Endophytic Bacteria for PGP traits
The potential of endophytic bacterial isolates for PGP activities were checked. Depending upon the extent of Phosphate solubilization, isolate OF115 was the highest phosphate solubilizer (450%). It is followed by isolate, WC138 (308%) as a good phosphate solubilizer, while WC2 (144%) was moderate phosphate solubilizer. On the other hand, all the tested endophytic isolates were found positive for IAA production (Appendices 3). Among all isolates, WC138 produced significant amount (10.809 μg/ml) of IAA followed by WC2 (10.571 μg/ml), OF115 (9.6 μg/ml) and WC58 (9.976 μg/ml). Moreover, all four isolates were able to produce ammonia (Appendices 3). These traits have been illustrated in Table 1.
Screening of isolates for Bio-control Activity
All isolates, except OF115 were found positive for their HCN production. The antagonistic activity of endophytic isolates against phytopathogenic fungi revealed the maximum antifungal zone for isolate, WC2 (32 mm) which was followed by OF115 (23 mm). On the other hand, the minimum antifungal zone was 11 mm produced by isolate, WC138 as shown in Table 1. Protease test was positive for WC58 only, while the rest of the strains failed to secrete this enzyme (Table 2).
| Isolated representative strains | F34 | F157 |
|---|---|---|
| Protease production | - | + |
| Amylase production | - | + |
| QTS tests | ||
| ONPG | + | + |
| CIT | + | + |
| MALO | + | + |
| LDC | + | + |
| ADH | + | + |
| ODC | + | + |
| H2S | + | + |
| UREA | + | + |
| TDA | + | + |
| IND | - | - |
| VP | - | - |
| GEL | - | - |
| GLU | + | - |
| NO3/N2 | + | + |
| MALT | + | + |
| SUC | + | - |
| MANN | + | - |
| ARAB | + | - |
| RHAM | + | + |
| SORB | - | - |
| INOS | - | - |
| ADO | - | - |
| MEL | - | - |
| RAF | + | - |
Table 2: Biochemical characterization of isolated representative strains.
Screening of endophytic bacteria for enzymatic activity
Bacterial endophytes were screened for their production of extracellular enzymes. Catalase activity was detected in all bacterial isolates. The results of amylase activity of endophytic isolates revealed that only WC58 showed strong amylase activity. The rest of the isolates were unable to produce this enzyme (Table 2).
16s rDNA and Phylogenetic analysis of the selected sequences
16s rDNA gene sequences indicated that WC2 shared 99% homology with Enterobacter cloacaeT and WC58 showed 99% homology with Enterobacter dissolvensT. In addition, WC138 was 100% homologue to Enterobacter asbuiaeT and OF115 showed 100% identity to Cronobacter sakazakii. The results of the BLAST analysis have been summarized in Table 2. The phylogenetic tree (Figure 1) constructed using 16s rDNA gene sequence reveals that WC138 make a separate clade with Enterobacter asburiae (AB004744) when compared with type strains and reference strains of closely related species, whereas WC2 is closely related to Enterobacter cloacae (HE978272) but also with Enterobacter dissolvens (Z96079) which is a subspecies of Enterobacter cloacae (HE978272), WC58 phylogenetically related to Enterobacter dissolvens (Z96079) while OF115 by making a separate clade is closely related to Cronobacter sakazakii (EF059843). The Sequence was submitted and the gene bank number were acquired from NCBI (Table 2).
Discussion
The plant endo-rhizosphere niche comprises a diverse population of bacterial taxa (endophytic bacteria) which may detrimental or beneficial to the associated plant. The bacterial endophytes isolated from traditional medicinal plants (W. coagulans and O. ferruginea) were investigated in the present study. The plants under investigation have been used as traditional mendicant’s to cure intestinal infections, asthma, biliousness, anti-tumor, anti-inflammatory, anti-stress, antioxidant, immunomodulatory, homeopathic and cardioprotective activities. The selected plants were found to harbor a variety of endophytic bacteria with varying population density among roots, leaves and stem of both plants. The numbers of endophytic isolates were greater in roots as compared to leaves and stems. It is a common trend that bacterial populations are larger in roots due to rhizosphere association, while less in stems and leaves as studied by [27]. The fact is that the soil serves as a main inoculum of microbe, so the roots being in close proximity with soil harbor a more diverse population of endophytic bacteria [28].
The phylogenetic tree constructed using 16s rDNA gene sequence revealed that the isolated endophytes belonging to diverse bacterial groups distributed in the family Enterobacteriacae. They were dominated by the genus Enterobacter followed by Cronobacter. To the best of our knowledge, both are being reported for the first time as an endophytic association with these medicinal plants i.e O. ferruginea and W. coagulans. The dominant existence and distribution of the genus Enterobacter with medicinal plants is in contrast to the result of earlier studies. Mostly, endophytic bacteria isolated from medicinal plants belonged to Pseudomonas and Bacillus species [29].
The genus Enterobacter has been extensively studied and reported to be endophytic colonizing hosts such as soybean, Citrus sinensis and crop plants [30]. Cronobacter has also been previously isolated from herbal tea plants [31], infant food, infant milk and cereal crops [32]. In addition, it has also been isolated from rhizosphere soil of plants growing in semi-arid region [33]. In four identified species E. cloacae and E. asburiae are commonly studied species and have been previously isolated as endophytes from Piper nigrum, corn, cotton, cucumber, rice, common bean [34-37]. However the data on the other two species is limited.
Nowadays interest is growing in exploring the endophyte potential for their plant growth promoting attributes and their usage as a biological control agent of plant pathogen. Functional diversity among all the isolates were observed for these traits. Phosphorus is needed by plants for growth and development, but its availability in agriculture soils is low [17]. Endophytes are known to improve plant growth by the phosphate solubilization [38]. In the present study, all strains were found to have the potential to solubilize phosphorous. As reported, the ability of endophytic Enterobacter species to solubilize phosphate on a large scale. The present study revealed E. asburiae as the potential plant growth promoting bacteria with the highest phosphate solubilizing efficiency, which can be related to an earlier study of who reported the same activity [39,40]. E. cloacae and E. dissolvens also solubilized phosphate and these are in accordance with the findings of [37,41,42]. While Cronobacter sakazakii has not been reported as phosphate solubilizer. It is reported that endophytes solubilize the inorganic phosphorus by reducing the pH through the excretion of organic acid activity, while organic phosphorous is solubilized by production of various phosphatases, which results in a better plant development and improved yield [43].
Production of IAA by endophytes is another valuable trait that influences plant growth directly [44]. The results of present strains with a significant amount of IAA production are similar to investigations performed by [37,40]. It has been reported that many endophytes including Enterobacter, Azotobacter, Serratia, Klebsiella produced IAA which stimulated plant growth [45]. In addition to IAA, ammonia production is another PGP trait which has a signaling role between plant and bacterial interactions [40]. Production of ammonia can be taken up by plants as a source of nitrogen [41]. Here it was observed that the most prominent activity was exhibited by E. asburiae which has also been reported by [40]. Crononbacter sakazakii has not been reported by any researcher for ammonia production.
Plant growth promoting bacteria have attracted much attention as biological control agents for their role in combating plant diseases, though their full potential has not yet been reached. Therefore, many of the bacterial endophytes promote plant growth indirectly by inhibiting the growth and activities of phytopathogens by the production of antimicrobial substances like HCN through a variety of different mechanisms. In the current study, HCN activity was displayed by E. cloacae and E. asburiae which are in line with the findings of other researcher [40,41]. It was observed that E. dissolves failed to produce HCN, which has also been reported by [46]. Crononbacter sakazakii is still not studied by previous worker for the production of HCN, because they were mostly isolated from non-living products. Moreover, the antagonistic ability of the bacterial endophytes has been evaluated. Enterobacter species towards Macrophomina phaseolina and other pathogenic fungi has been formerly tested by [47]. All the identified strains were for the first time checked against Macrophomina phaseolina which found to inhibit the growth of fungus by making large inhibition zones. The formation of antifungal zone may be due to the secretion of colicins by these bacteria that result in mycelial deformities.
Plant roots secreted numerous compounds which act as chemical attractants for a vast number of microbial communities. These exudates promote plant microbial beneficial symbiotic interactions [48,49]. Endophytic bacteria show more plant growth promoting effects than bacteria found in the rhizosphere [50]. It has been demonstrated that the isolated bacteria showed beneficial effects, such as secretion of growth promoting substances i.e. IAA, ammonia, phosphate solubilization. In addition, the isolated bacteria also act as biocontrol agent because they produced HCN and having antifungal activity which can inhibit phytopathogen. Protease is hydrolytic enzyme which is involved in the suppression of pathogenic growth and subsequent reduction in damage to plants [51]. Enterobacter dissolvens was able to produce this enzyme. This may be involved in the penetration of endophytes in host plant as described for Azoarcus sp.
Bacteria produce amylase to the outside of their cells to carry out extra cellular digestion [52]. In our present analysis, extracellular amylase enzymes were produced only by Enterobacter dissolvens. Amylase, which promotes early germination and increase in availability of starch assimilation [53]. On the other hand, the catalase enzyme is produced by the bacteria that respire using oxygen and protect them from the toxic by product of oxygen metabolism [54] and making them highly resistant to environmental, mechanical and chemical stresses [55]. All the bacterial strains show positive results for catalase activity.
Additionally, the tolerance of bacteria in stress conditions were determined. These properties may be helping host plants living in stress conditions by interactions of bacteria and plants. In the present study, the maximum salt tolerance of endophytic bacteria was observed at 2.5% and 7.5% salt concentration. It is noteworthy here that that the growth rate decreased with the increased in salt concentration. All bacterial isolates were tolerant with 2.5% sodium chloride, while none of the isolate tolerated 15% salt concentration. E. cloacae showed a high salt tolerance rate with a growth at 10% salt concentration, while the other two showed good growth at 7.5%, which is similar to previous studies [56]. The highly salt tolerant bacteria along with plant growth promoting traits would be advantageous for use in the mitigation of salt stress to make cultivation possible in saline agriculture lands [56].
Collective summarized analysis of all the experiments done in the present study highlighted the evident variation in the genetic makeup of the isolated endophytic bacterial strains which could be a valuable resource for applied agriculture. The information acquired from these strains could open the door and exploit the valuable data for the benefit of mankind. It is concluded that the isolated bacteria have numerous plant growth promoting activities which directly or indirectly promote plant growth.
References
- Dudeja S, Giri R, Saini R, et al. Interaction of endophytic microbes with legumes. J Basic Microbiol. 2012;52:248-60.
- Kobayashi DY, Palumbo JD. Bacterial endophytes and their effects on plants and uses in agriculture. Microbial Endophytes. 2000;19:199-233.
- Posada F, Vega FE. Establishment of the fungal entomopathogen Beauveria bassiana (Ascomycota: Hypocreales) as an endophyte in cocoa seedlings (Theobroma cacao). Mycologia. 2005;97:1195-200.
- Rosenblueth M, Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol Plant Microbe In. 2006;19:827-37.
- Tan Z, Hurek T, Gyaneshwar P, et al. Novel Endophytes of Rice form a Taxonomically Distinct Subgroup of Serratia marcescens. Syst Appl Microbiol. 2001;24:245-51.
- Zahran HH. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol Mol Bio Rev. 1999;63:968-89.
- Ullah A, Heng S, Munis MFH, et al. Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria: a review. Environ Exp Bot. 2015;117:28-40.
- Latif S, Khan S, Naveed M, et al. The diversity of Rhizobia, Sinorhizobia and novel non-Rhizobial Paenibacillus nodulating wild herbaceous legumes. Arch Microbiol 2013;195:647-53.
- Glick BR. The enhancement of plant growth by free-living bacteria. Canadian J Microbiol. 1995;41:109-17.
- Tudzynski B, Sharon A. Biosynthesis, biological role and application of fungal phytohormones. In: Industrial Applications. Springer. 2002;pp.183-211.
- Caldwell BA, Jumpponen A, Trappe JM. Utilization of major detrital substrates by dark-septate, root endophytes. Mycologia. 2000:230-32.
- Barrow J. Atypical morphology of dark septate fungal root endophytes of Bouteloua in arid southwestern USA rangelands. Mycorrhiza. 2003;13:239-47.
- Patten CL, Glick BR. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol. 2002;68:3795-801.
- Sergeeva E, Hirkala DL, Nelson LM. Production of indole-3-acetic acid, aromatic amino acid aminotransferase activities and plant growth promotion by Pantoea agglomerans rhizosphere isolates. Plant Soil. 2007;297:1-13.
- Mergeay M, Nies D, Schlegel H, et al. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162:328-34.
- Kiredjian M, Holmes B, Kersters K, et al. Alcaligenes piechaudii, a new species from human clinical specimens and the environment. Int J Sys Bacteriol. 1986;36:282-87.
- Pikovskaya R. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya. 1948;17:362-70.
- Qureshi M, Ahmad Z, Akhtar N, et al. Role of phosphate solubilizing bacteria (PSB) in enhancing P availability and promoting cotton growth. J Animal Plant Sci. 2012;22:204-10.
- Nguyen C, Yan W, Le Tacon F, et al. Genetic variability of phosphate solubilizing activity by monocaryotic and dicaryotic mycelia of the ectomycorrhizal fungus Laccaria bicolor (Maire) PD Orton. Plant Soil. 1992;143:193-9.
- Gordon SA, Weber RP. Colorimetric Estimation of Indoleacetic Acid. Plant Physiol. 1951;26:192-5.
- Cappuccino J, Sherman N. Negative staining. Microbiol Lab Manual. 1992:27-8.
- Lorck H. Production of hydrocyanic acid by bacteria. Physiol Plantarum. 1948;1:142-6.
- Sessitsch A, Reiter B, Berg G. Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can J Microbiol 2004;50:239-49.
- Sahu MK, Sivakumar K, Kannan L. Degradation of organic matters by the extra-cellular enzymes of actinomycetes isolated from the sediments and molluscs of the Vellar estuary. J Aqua Biol. 2005;20:142-4.
- Pearson WR. Flexible sequence similarity searching with the FASTA3 program package. In: Bioinformatics methods and protocols. 2000;132:185-219.
- Van de Peer Y, De Wachter R. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci. 1994;10:569-70.
- Rosenblueth M, Martínez-Romero E. Rhizobium etli maize populations and their competitiveness for root colonization. Arch Microbiol. 2004;181:337-44.
- Prakamhang J, Minamisawa K, Teamtaisong K, et al. The communities of endophytic diazotrophic bacteria in cultivated rice (Oryza sativa L). Appl Soil Ecol. 2009;42:141-9.
- Niranjanraj S, Deepak SA, Amruthesh KN, et al. Endophytic bacteria from different plant origin enhance growth and induce downy mildew resistance in pearl millet. Asian J Plant Pathol. 2007;1:1-11.
- Zinniel DK, Lambrecht P, Harris NB, et al. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol. 2002;68:2198-208.
- Stojanovic MM, Katic V, Kuzmanovic J. Isolation of Cronobacter sakazakii from different herbal teas. Vojnosanitetski Pregled. 2011;68:837-41.
- Shaker R, Osaili T, Al-Omary W, et al. Isolation of Enterobacter sakazakii and other Enterobacter sp. from food and food production environments. Food Control. 2007;18:1241-5.
- Bhatt PV, Vyas BRM. Screening and Characterization of Plant Growth and Health Promoting Rhizobacteria. Int J Curr Microbiol App Sci. 2014;3:139-55.
- Mcinroy JA, Kloepper JW. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil. 1995;173:337-42.
- Elbeltagy A, Nishioka K, Sato T, et al. Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Appl Environ Microbiol. 2001;67:5285-93.
- Hinton DM, Bacon CW. Enterobacter cloacae is an endophytic symbiont of corn. Mycopathologia. 1995;129:117-25.
- Jasim B, Jimtha CJ, Jyothis M, et al. Plant growth promoting potential of endophytic bacteria isolated from Piper nigrum. Plant Growth Reg. 2013;71:1-11.
- Schachtman DP, Reid RJ, Ayling SM. Phosphorus Uptake by Plants: From Soil to Cell Plant Physiol. 1998;116:447-53.
- Lopez BR, Bashan Y, Bacilio M. Endophytic bacteria of Mammillaria fraileana, an endemic rock-colonizing cactus of the southern Sonoran Desert. Arch Microbiol. 2011;193:527-41.
- Ahemad M, Khan MS. Plant growth promoting activities of phosphate-solubilizing Enterobacter asburiae as influenced by fungicides. Eur Asia J BioSci 2010;4:88-95.
- Deepa C, Dastager SG, Pandey A. Isolation and characterization of plant growth promoting bacteria from non-rhizospheric soil and their effect on cowpea (Vigna unguiculata (L.) Walp.) seedling growth. World J Microbiol Biotechnol. 2010;26:1233-40.
- Ramesh A, Sharma SK, Sharma MP, et al. Plant Growth-Promoting Traits in Enterobacter cloacae subsp. dissolvens MDSR9 Isolated from Soybean Rhizosphere and its Impact on Growth and Nutrition of Soybean and Wheat Upon Inoculation. Agrl Res. 2014;3:53-66.
- Rodri´guez H, Fraga R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol Adv. 1999;17:319-39.
- Shi Y, Lou K, Li C. Isolation, quantity distribution and characterization of endophytic microorganisms within sugar beet. African J Biotechnol. 2009;8.
- Spaepen S, Vanderleyden J, Remans R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS. 2007;31:425-48.
- Mehnaz S, Kowalik T, Reynolds B, et al. Growth promoting effects of corn (Zea mays) bacterial isolates under greenhouse and field conditions. Soil Biol Biochem. 2010;42:1848-56.
- Rani MU, Reddy G. Bacillus cereus and Enterobacter cancerogenus screened for their efficient plant growth promoting traits rhizobacteria (PGPR) and antagonistic traits among sixteen bacterial isolates from rhizospheric soils of Pigeon Pea. African J Microbiol Res. 2011;5:2090-4.
- Ahemad M, Kibret M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J King Saud Univ Sci. 2014;26:1-20.
- Ahmad E, Khan MS, Zaidi A. ACC deaminase producing Pseudomonas putida strain PSE3 and Rhizobium leguminosarum strain RP2 in synergism improves growth, nodulation and yield of pea grown in alluvial soils. Symbiosis. 2013;61:93-104.
- Dawwam G, Elbeltagy A, Emara H, et al. Beneficial effect of plant growth promoting bacteria isolated from the roots of potato plant. Ann Agr Sci. 2013;58:195-201.
- Bashan Y, de Bashan L. Plant growth-promoting. Enc Soils Environ. 2005;1:103-15.
- Poli A, Esposito E, Lama L, et al. Anoxybacillus amylolyticus sp. nov., a thermophilic amylase producing bacterium isolated from Mount Rittmann (Antarctica). Sys Appl Microbiol. 2006;29:300-07.
- Gholami A, Shahsavani S, Nezarat S. The effect of plant growth promoting rhizobacteria (PGPR) on germination, seedling growth and yield of maize. Int J Biol Life Sci. 2009;1:35-40.
- Ammor MS, Mayo B. Selection criteria for lactic acid bacteria to be used as functional starter cultures in dry sausage production: An update. Meat Sci. 2007;76:138-46.
- Kumar P, Khare S, Dubey R. Diversity of bacilli from disease suppressive soil and their role in plant growth promotion and yield enhancement. New York Sci J. 2012;5:90-111.
- Egamberdieva D, Kamilova F, Validov S, et al. High incidence of plant growth-stimulating bacteria associated with the rhizosphere of wheat grown on salinated soil in Uzbekistan. Environ Microbiol. 2008;10:1-9.