Research Article - Research and Reports on Genetics (2019) Volume 3, Issue 2
Prediction of circulating miRNAs and their target genes involved in venous thrombo-embolism (VTE) pathogenesis: A computational approach.
Srivastava S*, Garg I, Ganju L, Kumar B
Defence Institute of Physiology and Allied Sciences (DIPAS), Defence Research and Development Organization (DRDO), Timarpur, Delhi, India
- *Corresponding Author:
- Swati Srivastava
Defence Institute of Physiology and allied Sciences (DIPAS)
Defence Research and Development Organization (DRDO)
Delhi-110054
India
Tel: +919911135437
E-mail: sri_swati@rediffmail.com
Accepted Date: August 17, 2018
Citation: Srivastava S, Garg I, Ganju L, et al. Prediction of circulating miRNAs and their target genes involved in venous thrombo-embolism (VTE) pathogenesis: A computational approach. J Res Rep Genet. 2018;2(3):10-20.
Abstract
Venous thromboembolism (VTE) encompasses two clinically interrelated conditions; deep vein thrombosis (DVT) and pulmonary embolism (PE), the later one being potentially fatal. It is complex multifactorial disease which requires comprehensive understanding at molecular level. Currently, D-dimer is being clinically used for VTE diagnosis; however it has low specificity and diagnostic value. Till date, various experimental reports have discussed the functional role of microRNAs (miRNAs) in cardiovascular diseases, cancer, inflammation, atherosclerosis etc. Several recent studies have illustrated differential expression of various circulating miRNAs and their potential diagnostic and therapeutic values for VTE patients. These small RNAs are noncoding and suppress their gene targets at the post-transcriptional level. In the present study, we systematically evaluated available researches and performed bio-informatics analysis to provide new insights into the role of miRNA in VTE pathophysiology. Purpose of study: The aim of the present study was to consolidate the current information available on miRNAs linked to venous thrombosis and to identify the most potent miRNA and its target genes which could be useful in designing the future therapeutic strategies for treatment of venous thrombosis. Method: In the present study we retrieved recent articles associating various miRNAs with VTE pathophysiology. The eligibility criteria for inclusion in this study was met by six articles which pointed towards sixteen different miRNAs linked with VTE susceptibility. We further listed out their target genes using online miRNA target prediction tools and subjected each of them to functional gene enrichment analysis and pathway analysis. Results: Eleven functionally relevant miRNAs were identified after pathway analysis which regulate target genes involved in VTE associated pathways, such as hemostasis, blood coagulation, platelet activation, endothelin signalling and angiogenesis pathways. Other five miRNAs did not show any target genes in blood coagulation pathway and platelet activation, hence they were not considered for downstream analysis. The selected list of miRNAs was subjected to network analysis. Conclusions: Our in silico analysis results pointed towards four most potential miRNAs that could be used as biomarkers of VTE; hsa-mir-320a, hsa-mir-195, hsa-mir-103a-3p, hsa-mir- 26a. These miRNAs could also prove as critical targets for understanding molecular regulatory mechanism underlying pathophysiology and therapeutic interventions during VTE treatment. This analysis advance our understanding of regulatory mechanism of genes involved in VTE. Further in vivo validation of this data is required for confirming the present observation.
Keywords
Venous thrombosis, Deep vein thrombosis (DVT), Pulmonary embolism (PE), miRNA, Biomarkers, Hemostasis, Coagulation.
Abbreviation
VTE: Venous Thrombo-Embolism; DVT: Deep Vein Thrombosis; PE: Pulmonary Embolism; miR: Micro RNA; GO: Gene Ontology
Introduction
Venous thrombo-embolism (VTE) is a multi-factorial disease involving various genes and systems. Its two main clinical formations include DVT and PE, the later being a life-threatening complication. Both clinical forms of VTE are significant public concern as are regarded as the third most common vascular disease in developed countries [1] and is major cause of mortality and morbidity. It often occurs spontaneously and is asymptomatic, with no obvious clinical symptoms in early stages. Effective and timely therapy is needed in case of DVT, failing which abnormal swelling and ulceration of limbs occur and it may further lead to PE [2]. The risk factors of this complex disease, VTE, can be divided into acquired and genetic; the classic risk factors include cancer, surgery, prolonged immobilization, paralysis or fracture, hormone replacement therapy, use of oral contraceptive and hereditary coagulopathies. Blood clotting factors play a pivotal role in VTE pathogenesis. Increase in plasma levels of fibrinogen, factor VIII (FVIII), factor IX (FIX), factor XI (FXI) and prothrombin (FII) are considered as independent risk factors for VTE [3]. Also, since endothelium plays a key role in maintaining vascular homeostasis and controls coagulation, fibrinolysis and platelet activation [4-6], endothelial damage contributing to spontaneous and acute venous thrombosis has been well established [7-9]. Large numbers of laboratories all over the world are working on biomarkers that can be reliably used for VTE diagnosis. D-dimer test, which is currently used for VTE diagnosis has low specificity, as D-dimer levels are also elevated in non-thrombotic conditions such as infection, pregnancy, post-surgery and stroke [10]. Various types of venous malformations can be classified based on their severity and accurately diagnosed using different imaging techniques like contrast-enhanced MR-angiography [11,12]. Different radiological imaging patterns of different venous formations can help in deciding the treatment strategies [13]. However, these imaging techniques are costlier and are not available readily at all places. Therefore, despite vast research, consistent and reliable VTE biomarkers and specific gene targets for effective treatment are lacking.
Small RNA sequences such as Micro RNAs (miRNAs) form a part of evolutionarily conserved system. These small RNA sequences play a significant role in regulation of gene expression in eukaryotes. MiRNAs are small endogenous non-coding RNAs (~approx. 22 nucleotides) that suppress the expression of their target genes at post-transcriptional level through noncomplimentary base pairing with target mRNAs [14]. Each miRNA can potentially target several mRNAs and repress their activity thereby altering biological networks [15,16]. Although miRNAs act at intracellular level, circulating miRNAs can be detected in whole blood and plasma samples. Many strong evidences have been accumulated in the recent years which demonstrate the significant role of these miRNAs in pathogenesis of numerous cardiovascular diseases [17]. Several miRNAs have been shown to regulate the processes such as vascular inflammation, neointimal lesion formation, atherosclerosis and coronary artery disease [18-20]. Recent studies have reported differential expression of miRNA in venous thrombosis [21- 24]. Gradually, differentially expressed miRNAs in venous thrombosis are receiving attention. These VTE linked miRNAs can prove as potential biomarker candidates and also their target genes can enlighten pathogenetic mechanism and therapeutic options for VTE.
In the present study, we have systematically evaluated recent researches on miRNA linked to VTE/DVT and using various bioinformatic analysis tools further we enlisted most potential miRNAs and their target genes in this regard. Herein we provide new insights into the underlying mechanisms of VTE, for its diagnosis and therapeutics using targeted miRNA approach. Our findings advance understanding of regulatory mechanisms underlying VTE.
Methodology
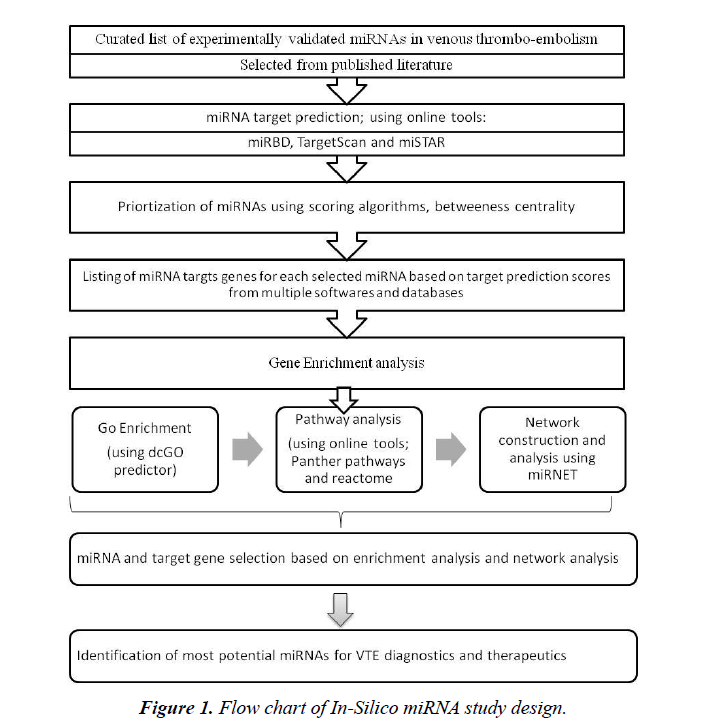
This study is based on review of published literature and further in silico analysis (Figure 1). Individual patient data is not involved in the present study and hence no ethical approval was necessary for this study.
Data sources and retrieval
Exhaustive web surfing was conducted for research articles published in databases PubMed, Embase and Web of Science in the period between 2000 to 2018. Based on this, a curated list of miRNAs with obvious role in venous thrombosis was prepared.
Inclusion criteria and data collection
Various articles relating miRNAs expression with DVT/VTE were contained. In the present in silico study, we have selected only those studies, wherein plasma or serum samples (peripheral source) were used to identify differentially expressed miRNAs; whereas those, in which tissue or cell lines were used as samples, were excluded from our study. Using the said selection criteria we aimed to study the potential use of circulating miRNAs in DVT diagnosis and treatment. Significantly expressed miRNAs from all these studies were selected for subsequent analysis (Table 1).
Table 1. List of selected miRNAs involved in VTE pathogenesis (based on published literature search).
| S. No. | Relevant miRNAs for VTE pathogenesis | Source | Regulation during VTE |
|---|---|---|---|
| 1 | hsa-miRNA-145 | [24] | Down-regulated in patients |
| 2 | hsa-miRNA-582 | [21] | Up-regulated in patients |
| 3 | hsa-miRNA-195 | [21] | Up-regulated in patients |
| 4 | hsa-miRNA-532 | [21] | Up-regulated in patients |
| 5 | hsa-miRNA-135 | [36] | Up-regulated in patients |
| 6 | hsa-miRNA-10b-5p | [22] | Up-regulated in patients |
| 7 | hsa-miRNA-320a | [22] | Up-regulated in patients |
| 8 | hsa-miRNA-320b | [22] | Up-regulated in patients |
| 9 | hsa-miRNA-424-5p | [22] | Up-regulated in patients |
| 10 | hsa-miRNA-423-5p | [22] | Up-regulated in patients |
| 11 | hsa-miRNA-103a-3p | [22] | Down-regulated in patients |
| 12 | hsa-miRNA-191-5p | [22] | Down-regulated in patients |
| 13 | hsa-miRNA-301a-3p | [22] | Down-regulated in patients |
| 14 | hsa-miRNA-199-3p | [22] | Down-regulated in patients |
| Repeat | hsa-miRNA-424-5p | [23] | Up-regulated in patients |
| 15 | hsa-miRNA-136-5p | [23] | Down-regulated in patients |
| 16 | hsa-miRNA-26a | [37] | Down-regulated in patients |
Target identification
Three different online tools, viz. Target scan (http://www.targetscan.org/vert_71/) [25], miRDB (http://www.mirdb.org/) [26] and miSTAR (www.mi-star.org) (miRNA stacked model target prediction) [27] were used to identify target genes of selected miRNA. These tools comprise of comprehensive data of experimentally validated miRNA-mRNA target interactions. Also, these databases are updated periodically to incorporate recent information. Based on these, miRNA target search was conducted and a list of target genes for each selected miRNA was prepared. For mRNA prediction, both predicted and experimentally validated targets were chosen from each database, based on the target prediction scores.
Functional gene annotation and pathway analysis
The target gene lists thus obtained were subjected to functional gene annotation using Gene Codis (http://genecodis.cnb.csic.es/analysis) [28-30]. GO (Gene Ontology) functional analysis, or “GO term enrichment,” was used to identify pathways and processes that are significantly enriched for each set of genes. Enrichment analysis was also used to identify pathways associated with target genes of miRNA. Pathway analysis was done using Reactome (https://reactome.org/PathwayBrowser/) [31,32] and Panther pathways (http://pantherdb.org/tools/index.jsp) [33].
Network analysis
All the sixteen miRNAs were subjected to computation based network analysis. Network building was done using web tool miRNet (http://www.mirnet.ca.) [34], which generated high quality miRNA-target interaction networks using various databases. Different filters were applied on miRNAs to make the networks more clear by controlling its size and not losing any important information. These filtering tools were based on topological measures such as degree, betweeness and shortest path. With this way, we could manage fine control over resulting networks.
Comparison of target genes
Target genes regulated by selected miRNAs were compared using Venn diagram analysis. Venn diagrams were obtained using http://www.interactivenn.net/ [35].
Results
miRNAs involved in VTE
All recent publications available online were put under scrutiny for initial selection of most potent miRNA in relation to VTE pathophysiology. Since the circulating miRNAs found in body fluids such as plasma or serum are apparently more stable, hence we selected only those studies wherein miRNAs extracted from peripheral source were taken. After screening, the studies conducted on certain tissues and cell lines were excluded and duplicates were removed. We selected 16 potentially relevant miRNAs based on results of six recent studies, which met the criteria [21-24,36,37] for further analysis. Both Qin et al. [21] and Wang et al. [23] pooled their samples to decrease redundancy amongst individuals.
Significant miRNA expression
The selected articles and their experimentally validated significant miRNAs associated with VTE/DVT are shown in Table 1. Notably, miR-424-5p was the only common miRNA in two research publications. Out of sixteen miRNAs selected in total, nine were found to be up-regulated and seven were downregulated in VTE patients.
Identification of target genes of significant miRNAs
It is well established that each miRNA is able to regulate from one to a large number of mRNAs, different algorithms are available to search miRNA–target interactions. In order to make the method more robust, three different online software were used to identify target genes of selected differentially expressed miRNAs; miSTAR, miRBD and Target Scan. These genes were selected based on their target prediction score. After removing the duplicate genes from each list, the number of genes targeted by each selected miRNA was found to be highly variable (Table 2).
Table 2. Listing number of target genes for selected miRNAs based on target prediction score.
| S. No. | Relevant miRNAs for VTE pathogenesis | Target genes (miSTAR, miRDB, TargetScan) |
|---|---|---|
| 1 | hsa-miRNA-145 | 1456 |
| 2 | hsa-miRNA-582 | 1704 |
| 3 | hsa-miRNA-195 | 1091 |
| 4 | hsa-miRNA-532 | 1553 |
| 5 | hsa-miRNA-135 | 723 |
| 6 | hsa-miRNA-10b-5p | 142 |
| 7 | hsa-miRNA-320a | 482 |
| 8 | hsa-miRNA-320b | 343 |
| 9 | hsa-miRNA-424-5p | 763 |
| 10 | hsa-miRNA-423-5p | 361 |
| 11 | hsa-miRNA-103a-3p | 739 |
| 12 | hsa-miRNA-191-5p | 70 |
| 13 | hsa-miRNA-301a-3p | 760 |
| 14 | hsa-miRNA-199-3p | 407 |
| 15 | hsa-miRNA-136-5p | 303 |
| 16 | hsa-miRNA-26a | 1259 |
Functional classification of miRNAs
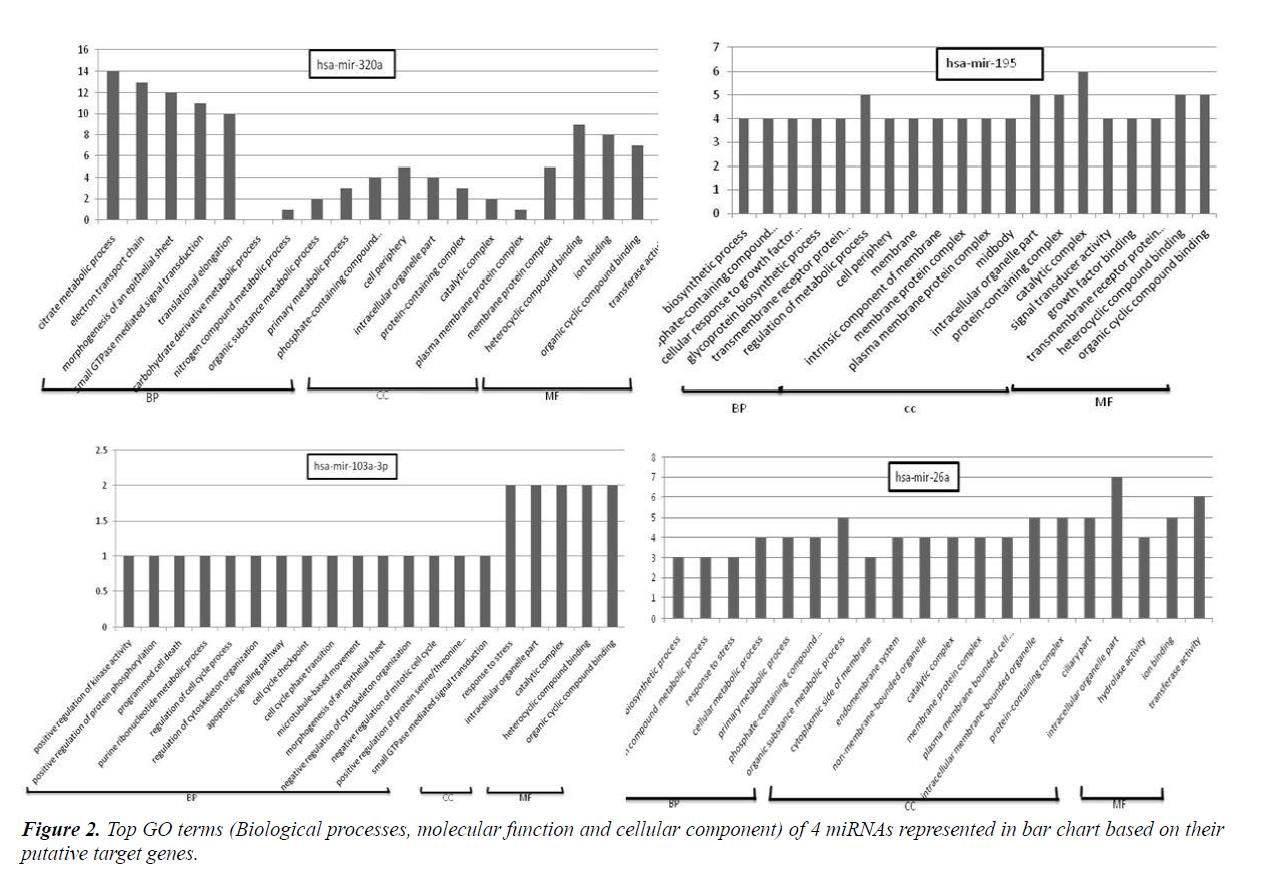
GO assigns biological processes (BP), cellular components (CC) and molecular function (MF). Online software dcGO predictor was used to perform GO classification (supfam.org/ SUPERFAMILY/cgi-bin/dcpredictormain.cgi). Statistical significant cut-off p-value was set as <0.05. GO enrichment analysis categorized miRNA target genes into biological processes, molecular function and cellular component genes. Amongst top terms of biological processes included those belonging to regulation of cell cycle process, programmed cell death, response to stress, apoptosis signalling pathway, positive regulation of protein serine/threonine, cellular metabolic process etc. Terms included in molecular function were those involved in protein binding, DNA binding, RNA binding, nucleoside phosphate binding, transferase activity, oxireductase activity etc. Meanwhile cell component terms were enriched in intracellular organelle, protein-containing complex, plasma membrane protein complex, cell periphery and catalytic complex. The representative GO terms for 4 miRNAs are shown in Figure 2.
Pathway analysis
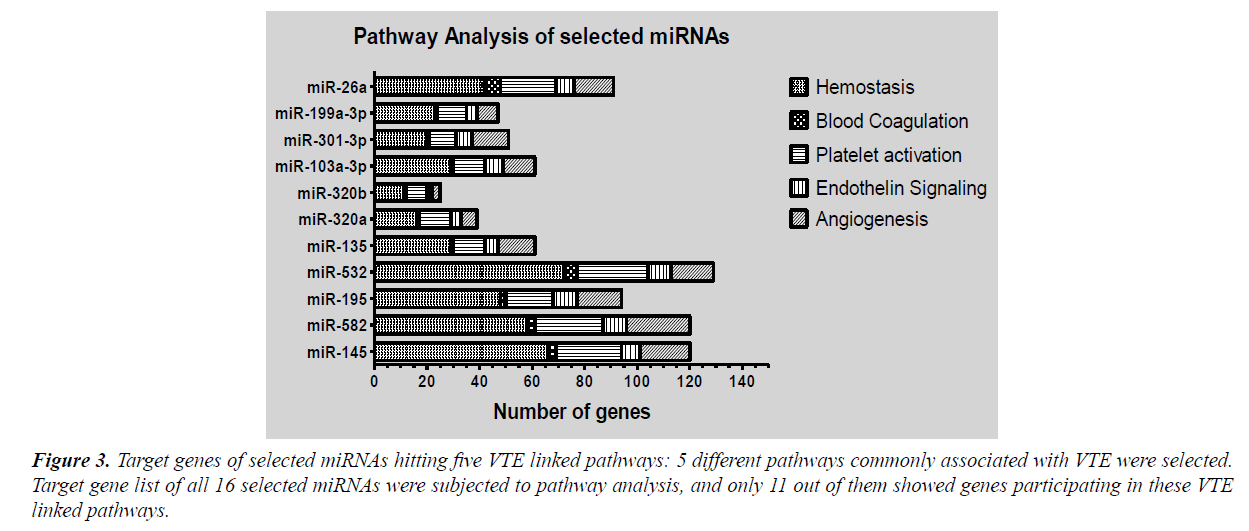
Pathway enrichment was done using Panther pathways and Reactome pathway database. Enrichment analysis enlisted large number of pathways regulated by target genes of significant miRNAs. These included various signalling pathways like mitogen activated protein kinase (MAPK) signalling, alpha adrenergic receptor signalling, apoptosis signalling, EGF receptor signalling, FGF signalling, interleukin signalling, Wnt signalling, Notch signalling and VEGF signalling along with genes of glycolysis pathway, RAS pathway, TCA cycle, DNA replication, oxidative stress response, T-cell activation pathway etc. In order to narrow the miRNA list and evaluate the role of selected miRNAs in VTE, we selected five pathways based on their established role in pathogenesis of venous thrombosis viz., hemostasis, blood coagulation, platelet activation endothelin signalling and angiogenesis. The extracted target genes for all selected miRNAs were checked for their role in mediating these pathways. Target genes of only eleven miRNAs out of sixteen were participating in the selected pathways (Figure 3). This converged our focus to eleven miRNAs instead of sixteen.
Figure 3: Target genes of selected miRNAs hitting five VTE linked pathways: 5 different pathways commonly associated with VTE were selected. Target gene list of all 16 selected miRNAs were subjected to pathway analysis, and only 11 out of them showed genes participating in these VTE linked pathways.
Network analysis
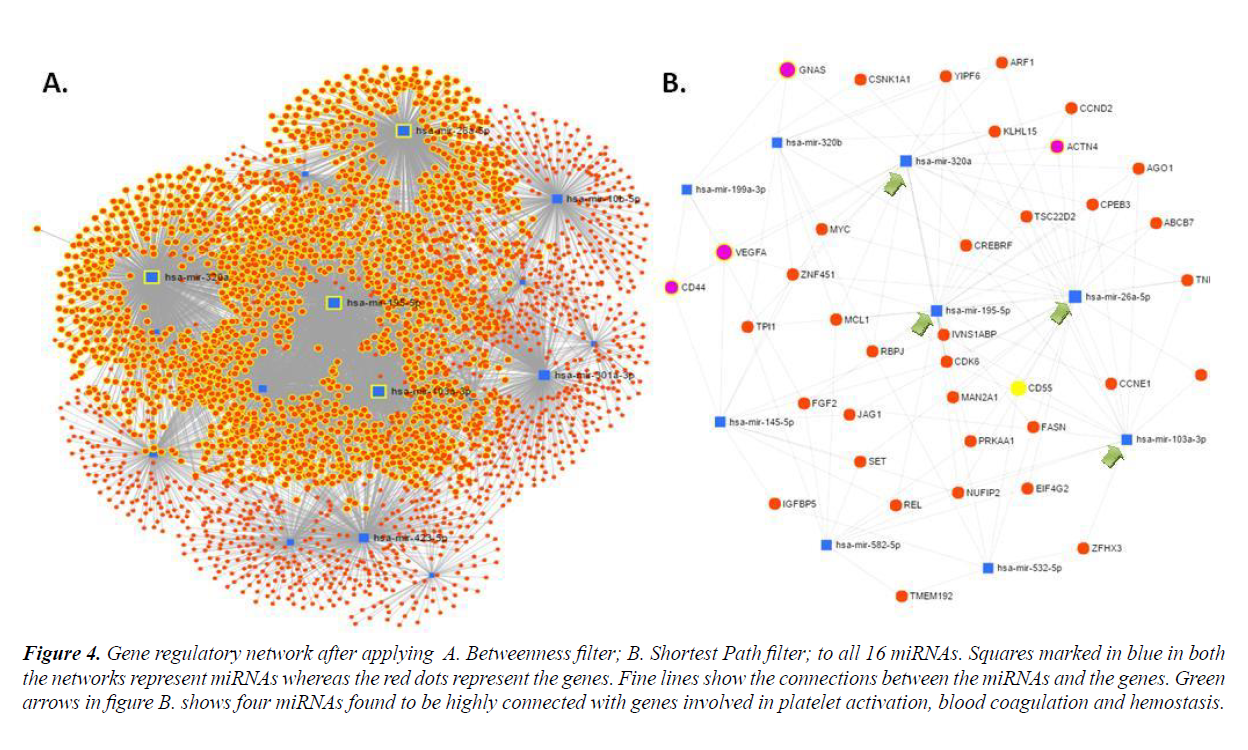
In order to establish molecular interactions of miRNA-target genes, miRNA mediated gene regulatory networks were constructed. Detailed study of the network properties was done such as betweenness centrality for the network, which measured the number of shortest paths going through the node taking into consideration the global network structure and composed of degree which took into consideration the minimal betweenness and was by default set as 0.0. All the sixteen selected miRNAs were subjected to network creation using miRNet. Networks showed varying degree and betweenness centrality amongst them, as shown in Table 3. MiRNA-target interaction networks were optimized using different algorithms and filters to improve visualization. Four miRNAs viz. hsa-miR-320a, hsa-miR- 199-5p, hsa-miR103a-3p and hsa-miR-26a-5p were found to show maximum nodes and were positioned between the dense clusters (Figure 4). These four miRNA species were found to be closely connected to genes involved in platelet activation, blood coagulation and hemostasis etc., as listed in Table 4.
Figure 4: Gene regulatory network after applying A. Betweenness filter; B. Shortest Path filter; to all 16 miRNAs. Squares marked in blue in both the networks represent miRNAs whereas the red dots represent the genes. Fine lines show the connections between the miRNAs and the genes. Green arrows in figure B. shows four miRNAs found to be highly connected with genes involved in platelet activation, blood coagulation and hemostasis.
Table 3. List of top miRNAs based on scoring algorithm.
| S.No. | miRNA | Degree | Betweenness centrality |
|---|---|---|---|
| 1 | hsa-mir-320a | 584 | 1329400 |
| 2 | hsa-mir-26a | 457 | 1048200 |
| 3 | hsa-mir-195-5p | 640 | 1034700 |
| 4 | hsa-mir-301a-3p | 395 | 910980 |
| 5 | hsa-mir-423-5p | 343 | 777670 |
| 6 | hsa-mir-103a-3p | 453 | 764230 |
| 7 | hsa-mir-10b-5p | 323 | 757830 |
| 8 | hsa-mir-145-5p | 238 | 528900 |
| 9 | hsa-mir-424-5p | 471 | 475930 |
| 10 | hsa-mir-136-5p | 160 | 358660 |
| 11 | hsa-mir-582-5p | 149 | 283790 |
| 12 | hsa-mir-199a-3p | 114 | 202370 |
| 13 | hsa-mir-135a-5p | 94 | 201440 |
| 14 | hsa-mir-191-5p | 78 | 164240 |
| 15 | hsa-mir-320b | 145 | 137070 |
| 16 | hsa-mir-532-5p | 64 | 131630 |
Table 4. List of genes from relevant pathways and their biological role, obtained from network analysis.
| S. No. | Biological role | Gene Count | Included gees |
|---|---|---|---|
| 1 | Platelet aggregation | 9 | AKT1, MPL, PTK2, SRC, ADRA2B, CRK, GRB2, ITGB3, SYK |
| 2 | Blood coagulation | 3 | ATP2B1, TRPC6, ITPR1 |
| 3 | Platelet Adhesion to exposed collagen | 1 | ITGA2 |
| 4 | Homeostasis | 8 | ATP2B1, NOS2, TRPC6, GNB1, MAPK14, GNAS, ITPR1, PTPN11 |
| 5 | eNOS activation / Endothelial response | 4 | CAV1, AKT1, CALM3, HSP90AA |
| 6 | Complement cascade | 2 | C3, CD55 |
| 7 | Cellular response to hypoxia | 8 | EP300, RPS27A, VEGFA, EPAS1, HIF1A, CUL2, UBA52, VHL |
| 8 | Cellular responses to stress | 88 | ATM, CDK6, E2F2, EP300, EZH2, GSK3B, HMGA1, HSPA8, IL6, MDM2, RPS27A, YWHAE, HMGA2, HIST1H4J, HIST1H4E, BAG4, PRDX3, NUPL2, TNRC6B, NUP205, AGO1, TNRC6A, EP400, HIST4H4, CDK2, NUP50, PHC3, AGO3, MAPK14, ETS2, MTOR, MAPK1, MAPK8, MAPK9, SOD2, VEGFA, E2F1, GPX8, CDK4, CDKN1A, E2F3, EPAS1, ETS1, HIF1A, MAP2K6, RPA1, RPS6KA3, MAP2K4, SP1, NUP43, CDC27, HSPA1, MAP2K3, CUL2, CBX4, MAPKAPK2, CBX6, MINK1, CBX2, AGO4, ATP7A, BMI1, CDKN2A, HSP90AB1, RELA, RPS6KA1, UBA52, HIST1H3H, HIST1H3B, PRDX6, POM121, SUZ12, CABIN1, TNRC6C, HIST2H3A, HIST2H4B, VHL, HSP90AA1, ERO1A, MAPK3, EHMT2. |
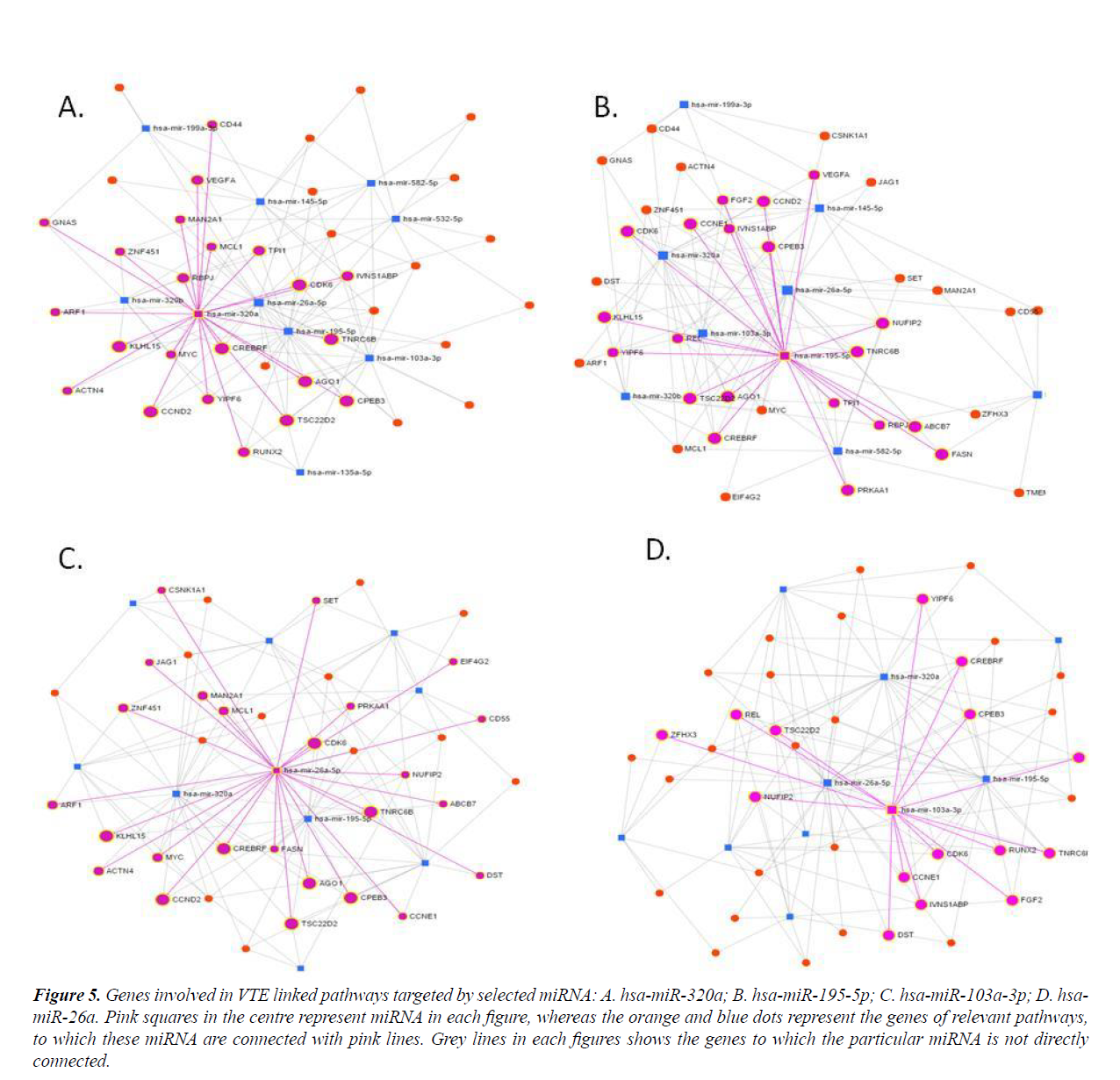
Many important genes involved in VTE relevant pathways are targeted by these four miRNAs (Figure 5). Thus miRNA-mRNA networks highlighted four central miRNAs which showed strongest interactions with the genes of interest.
Figure 5: Genes involved in VTE linked pathways targeted by selected miRNA: A. hsa-miR-320a; B. hsa-miR-195-5p; C. hsa-miR-103a-3p; D. hsamiR- 26a. Pink squares in the centre represent miRNA in each figure, whereas the orange and blue dots represent the genes of relevant pathways, to which these miRNA are connected with pink lines. Grey lines in each figures shows the genes to which the particular miRNA is not directly connected.
Comparison of target genes regulated by 4 selected miRNAs
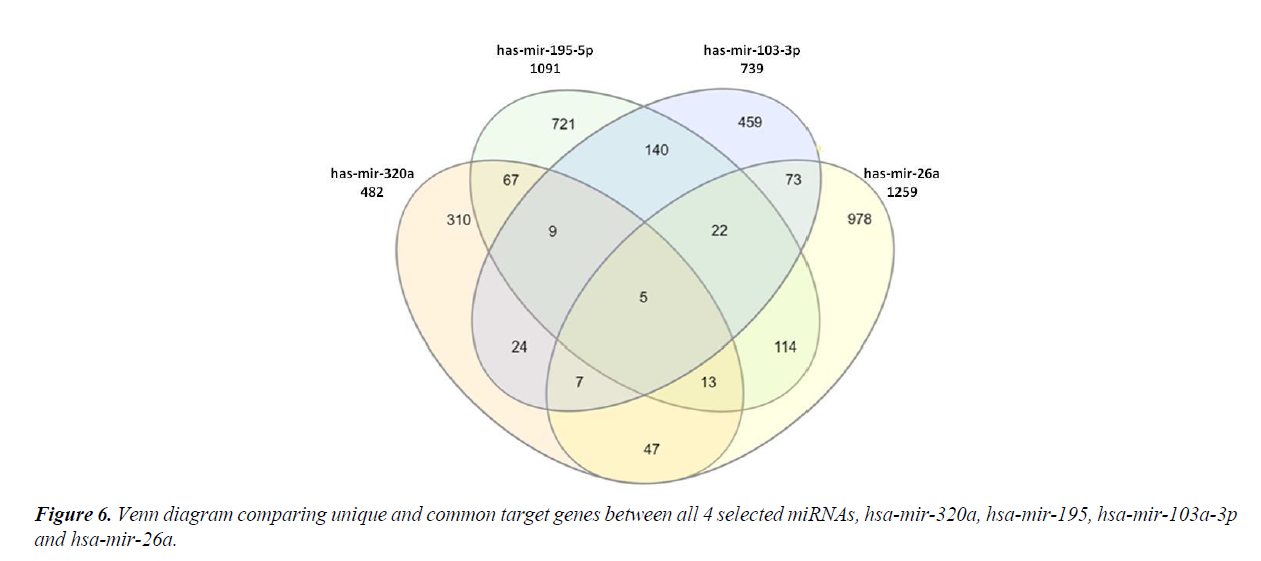
The four most relevant miRNAs obtained after network analysis were subjected to comparison amongst each other, by comparing the difference and similarities in their target genes. There were only five genes which were regulated by all the four miRNAs (Figure 6). These were CDK6, CELF2, CREBRF, DCP1A and PDIK1L. Also, a large number of genes were found to be uniquely targeted by each miRNA, viz., 310 genes by hsa-miR- 320a, 721 genes by hsa-miR-195-5p, 459 genes by hsa-mir- 103a-3p and 978 genes by hsa-miR-26a (Figure 6).
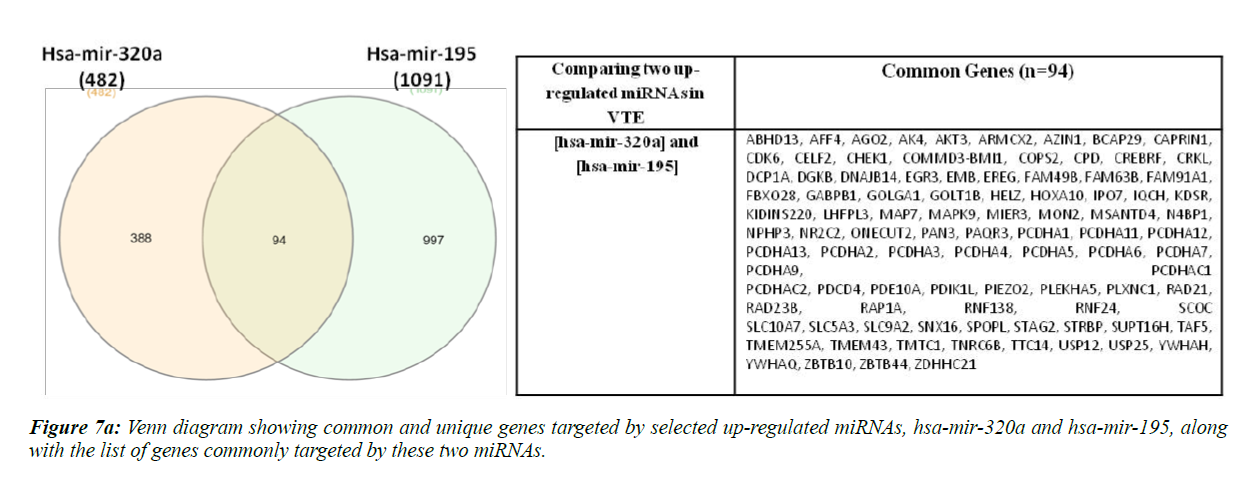
Hsa-miR-320a and hsa-mir-195-5p are reportedly up-regulated whereas hsa-miR-103a-3p and ha-mir-26a are down-regulated in VTE/DVT patients [21,22,37]. We compared the target gene list of these two up-regulated and down-regulated miRNAs separately, as shown in Figure 7a and 7b.
Ninety four genes were found to be commonly targeted upon comparison of hsa-mir-320a and hsa-mir-195. Sine these miRNAs have been found to be up-regulated in VTE/DVT patients, these genes should be down-regulated in such patients. Also, when we compared hsa-mir-103a-3p and hsa-mir-26a, 107 genes were found to be commonly regulated and since these miRNAs have been previously shown to be down-regulated in VTE/DVT, the genes regulated by these miRNAs should be upregulated in VTE patients.
Discussion
The term ‘hemostasis’ refers to maintaining the blood fluidity under various physiological conditions and sudden stoppage of blood flow in case of vascular injury to prevent haemorrhage in addition to inappropriate thrombus formation [38]. The most important players in maintaining hemostatic balance in the body are platelets, coagulation factors, fibrinolytic factors and vascular endothelium. The delicate interplay of all these ensure adequate blood flow. However, several intrinsic or extrinsic, acquired or genetic factors may lead to disruption this fine tuning resulting in imbalance of hemostatic system. This may further lead to hyper-coagulable state (thrombosis) or hypo-coagulable state (bleeding) [39]. Several studies have been conducted so far to understand the pathophysiology of venous thrombosis. Diagnostic marker most commonly used for DVT diagnosis, D-dimer, is highly sensitive although it lacks specificity, resulting in false positive results. Thus a large number of studies are focussing on discovery of alternative biomarkers for DVT like selectins, micro particles, IL-10 and other inflammatory markers [40]. With the advances in miRNA screening technologies, many studies have determined role of miRNAs in various pathological conditions, particularly cardiovascular diseases [41-43] and cancer [44,45]. MiRNAs represent a unique epigenetic mechanism that regulates gene expression in many homoeostatic processes and pathological conditions within the cells. Also, few recent studies have conducted an in-silico search and found that some predicted gene targets encoding for cell adhesion molecules, platelets and endothelial growth factors which could potentially result in prothrombotic conditions [22,23].
We performed in-silio computational analysis to investigate the involvement of previously reported miRNAs in VTE prognosis. Sixteen potential miRNAs were shortlisted for this purpose after extensive literature search. Interestingly, all included studies have shown varying miRNA profiles. Only miR-424-5p was identified in two independent studies [22,23]. Wang et al. showed up-regulation of miR-424-5p in plasma of DVT/VTE patients and its levels were significantly related to hypercoagulability indexes, D-dimer and APC-PCI values. However, our present in-silico analysis could not replicate link between miR-424-5p and VTE linked genes.
Recent evidences have revealed that circulating miRNAs, are important regulatory molecule. Many such regulatory miRNAs have emerged as a promising class of biomarkers in many cardiovascular diseases, malignancies as well as VTE. Thus we selected miRNAs which have been isolated from peripheral tissues, serum and plasma, and studied further. In silico approach is being widely used these days to identify most important pathways and genes regulated by miRNA signatures. Knowledge of target mRNAs of a particular miRNA is imperative to understand the role of particular a miRNA in both normal cellular processes and pathogenesis. Although VTE is a multi-factorial disease and has involvement of multiple genes and pathways, however pathways such as platelet activation and aggregation, hemostasis, blood coagulation and endothelial gene function pathways has direct involvement in VTE occurrence and pathogenesis [46-49]. We found that target genes of 11 out of sixteen miRNAs hit these VTE linked pathways. Whereas, none of the gene targets of hsa-miR-10b-5p, hsa-miR-424-5p, hsa-miR-423-5p, hsa-miR-191-5p and hsa-miR-136-5p were included in to blood coagulation pathways. Interestingly, hsamiR- 26a targets showed 6 genes of blood coagulation pathway, 21 genes of platelet activation and aggregation and 42 genes of hemostasis pathway, showing its functional relevance in VTE.
Functional annotation of miRNAs selected from pathway analysis, showed gene enrichment of many GO terms, most importantly including cell proliferation, response to stress, programmed cell death, cell division, cellular response to stimulus, cell signalling, blood vessel development, signal transduction, apoptotic signalling etc. Since endothelial cells play a significant role in thrombosis formation and hemostasis at different sites [49], the genes involved in endothelial cell proliferation, division, signalling and blood vessel development might be involved in overall vascular growth and repair and hence play an important feedback role in thrombosis stimulation.
Network analysis further revealed that four miRNAs viz., hsamiR- 320a, hsa-miR-199-5p, hsa-miR103a-3p and hsa-miR- 26a-5p have shown maximum connected nodes with platelet activation and hemostasis genes and thus could be strong predictors of VTE. These four miRNAs could be critical targets for understanding VTE pathophysiology and could also be used as potential therapeutic targets for treatment. Moreover, the genes commonly targeted by these miRNAs could be playing an important role in VTE development. Meanwhile, analysis of miRNA-target genes (Figure 7a and 7b) could enlighten new insights into the diagnosis and pathogenesis of VTE. Thus, in the present analysis, we present a comprehensive relationship between these four circulating miRNAs and VTE. Nevertheless, this relationship needs an experimental validation. Furthermore, there are certain inevitable challenges in this area of research like single miRNA may target hundreds of gene and similarly a single gene may be targeted by number of miRNAs [50,51].
Conclusion
Conventional biomarkers lack specificity for diagnosis and prognosis of venous thrombosis. Since current diagnostic and therapeutic strategies exhibit a poor performance and surveillance in case of VTE, there is an immediate need of reliable biomarker and therapeutic target for effective prevention and treatment of VTE. Recent work on micro RNAs suggest their important role in regulating complex biological processes and their dysregulation which can in turn lead to complex diseases and pathological conditions. Few recent studies have been done on role of miRNAs in VTE/DVT pathophysiology. Present in-silico computational analysis was planned to integrate the current information on miRNA discovery, target identification and elucidation of their regulatory mechanisms in relation to VTE. Results of our analysis revealed four miRNAs and their key target genes that could be associated with VTE and could prove as potential diagnostic markers and therapeutic targets for VTE in future. More in-depth and larger sample investigations in wet lab are needed to explore the diagnostic and therapeutic values of these miRNAs for VTE/DVT. Limitation: This is an insilico study based on available data and online tools for various analysis. The relevant miRNAs and genes identified in the study could be logically linked to VTE pathogenesis, however the data obtained here needs to be experimentally validated.
Acknowledgement
Authors are thankful to the lab staff of Genomics group for their support and cooperation during the study.
References
- Tapson VF, Humbert M. Incidence and prevalence of chronic thromboembolic pulmonary hypertension: From acute to chronic pulmonary embolism. Proc Am Thorac Soc. 2006;3:564-7.
- Wang W, Li C, Li W, et al. MiR-150 enhances the motility of EPCs in vitro and promotes EPCs homing and thrombus resolving in vivo. Thromb Res. 2014;133:590-8.
- Bertina RM. Elevated clotting factor levels and venous thrombosis. Pathophysiol Haemost Thromb. 2003;33:395-400.
- Levi M, Cate HT, Poll TV. Endothelium: interface between coagulation and inflammation. Crit Care Med. 2002;30:S220-S4.
- Stern DM, Esposito C, Gerlach H, et al. Endothelium and regulation of coagulation. Diabetes Care 1991;14(2):160-6.
- Reichenbach G, Momi S, Gresele P. Nitric oxide and its antithrombotic action in the cardiovascular system. Curr Drug Targets Cardiovasc Haematol Disord. 2005;5(1):65-74.
- Thomas DP, Merton RE, Wood RD, et al. The relationship between vessel wall injury and venous thrombosis: an experimental study. Br J Haematol. 1985;59(3):449-57.
- Migliacci R, Becattini C, Pesavento R, et al. Endothelial dysfunction in patients with spontaneous venous thromboembolism. Haematologica. 2007;92(6):812-8.
- Prasad M, McBane R, Reriani M, et al. Coronary endothelial dysfunction is associated with increased risk of venous thromboembolism. Thrombosis Research. 2016;139:17-21.
- Bates SM, Jaeschke R, Stevens SM, et al. Diagnosis of DVT: antithrombotic therapy and prevention of thrombosis, 9th ed: American College Chest Physicians Evidence-Based Clinical Practice Guidelines. Chest 2012;141:351S-418S.
- Razek AA, Alvarez H, Bagg S, et al. Imaging spectrum of CNS vasculitis. Radiographics. 2014;34:873-94.
- Razek AA, Saad E, Soliman N, et al. Assessment of vascular disorders of the upper extremity with contrast-enhanced magnetic resonance angiography: pictorial review. Jpn J Radiol. 2010;28:87-94.
- Razek AA, Ashmalla G, Samir S. Clinical value of classification of venous malformations with contrast enhanced MR angiography. Phlebology. 2017;32:628-33.
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610.
- Selbach M, Schwanhäusser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63.
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233.
- Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNA: novel biomarkers and extracellular communicators in cardiovascular diseases? Cir Res. 2012;110:483-95.
- Ji R, Cheng Y, Yue J, et al. Microrna expression signature and antisense-mediated depletion reveal an essential role of microrna in vascular neointimal lesion formation. Circ Res. 2007;100:1579-88.
- Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ. Res. 2009;104:476-87.
- Weber C., Schober A., Zernecke A. Micrornas in arterial remodelling, inflammation and atherosclerosis. Curr. Drug Targets. 2010;11:950-6.
- Qin J, Liang H, Shi D, et al. A panel of microRNAs as a new biomarkers for the detection of deep vein thrombosis. J Thromb Thrombolysis. 2015;39:215-21.
- Starikova I, Jamaly S, Sorrentino A, et al. Differential expression of plasma miRNAs in patients with unprovoked venous thromboembolism and healthy control individuals. Thromb Res. 2015; 136(3):566-72.
- Wang X, Sundquist K, Elf JL, et al. Diagnostic potential of plasma microRNA signatures in patients with deep vein thrombosis. Thromb Haemost. 2016;116: 328-36.
- Sahu A , Jha PK , Prabhakar A , et al. MicroRNA-145 Impedes Thrombus Formation via Targeting Tissue Factor in Venous Thrombosis. EBioMedicine. 2017;26:175-186.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4.
- Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015; 43:146-52.
- Peer VG, De Paepe A, Stock M, et al. miSTAR: miRNA target prediction through modeling quantitative and qualitative miRNA binding site information in a stacked model structure. Nucleic Acids Res. 2017;45(7).
- Madrid TD, Cadenas NR, Pascual-Montano A: GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012.
- Cadenas NR, Saez CP, Vazquez M, et al. GeneCodis: interpreting lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009.
- Saez CP, Chagoyen M, Triado F, et al. Genecodis: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8(1).
- Croft D, Mundo AF, Haw R, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:472-7.
- Fabregat A, Jupe S, Matthews L, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46:649-55.
- Mi H, Dong Q, Muruganujan A, et al. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:204-10.
- Fan Y, Siklenka K, Arora SK, et al. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;8:44:135-41.
- Heberle H, Meirelles GV, da Silva, et al. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16(1):169.
- Xiao J, Jing Z-C, Ellinor PT, et al. MicroRNA-134 as a potential plasma biomarker for the diagnosis of acute pulmonary embolism. Journal of Translational Medicine. 2011;9:159.
- Li Z, Ni J. Role of microRNA-26a in the diagnosis of lower extremity deep vein thrombosis in patients with bone trauma. Exp Ther Med. 2017;14(5):5069-74.
- Versteeg HH, Heemskerk JW, Levi M, et al. New fundamentals in hemostasis. Physiol Rev. 2013;93:327-58.
- Rasche H. Haemostasis and thrombosis: an overview. European Heart Journal Supplements. 2001;3:Q3-Q7.
- Coleman DM, Wakefield TW. Biomarkers for the diagnosis of deep vein thrombosis. Expert Opin Med Diagn. 2012;6(4):253-7.
- Yepes S, Lopez R, Andrade RE, et al. Co-expressed miRNAs in gastric adenocarcinoma. Genomics. 2016;108:93-101.
- Louwies T, Vuegen C, Panis LI, et al. miRNA expression profiles and retinal blood vessel calibers are associated with short-term particulate matter air pollution exposure. Environ Res. 2016;147:24-31.
- Murphy MS, Casselman RC, Tayade C, et al. Differential expression of plasma microRNA in preeclamptic patients at delivery and 1 year postpartum. Am J Obstet Gynecol. 2015;213:367.
- Kim H, Yang JM, Jin Y, et al. MicroRNA expression profiles and clinicopathological implications in lung adenocarcinoma according to EGFR, KRAS, and ALK status. Oncotarget. 2017;8(5):8484-98.
- Yan J, She Q, Shen X, et al. Potential Role of MicroRNA-375 as Biomarker in Human Cancers Detection: A Meta-Analysis. BioMed Research International. 2017.
- Garcia SM, Schindewolf M, Stanford S, et al. The Role of Platelets in Venous Thromboembolism. Semin Thromb Hemost. 2016;42(3):242-51.
- Tripodi. Levels of coagulation factors and venous thromboembolism. Haematologica. 2003;88(6):705-11.
- Rosendaal FR, Reitsma PH. Genetics of venous thrombosis. J Thromb Haemost. 2009;1:301-4.
- Kawaan HC, Samama MM. The significance of endothelial heterogeneity in thrombosis and hemostasis. Semin Thromb Haemost. 2010;36:286-300.
- Grimson A, Farh KK, Johnston WK et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mil Cell. 2007;27:91-105.
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are miroRNA targets. Cell. 2005;120:15-20.