Research Article - Journal of Clinical and Experimental Toxicology (2018) Volume 2, Issue 2
Novel benzoxazole derived genotoxicity apoptogenic effects in hepatoma cell.
Yaoqing Chen1*, Yang Wang2*#, Xueli Xie1, Abhay Pratap Singh3, Jian Fu1*1Department of Pharmacology, Hainan Medical College, Haikou, China
2Laboratory of Extreme Environment Sports Medicine and Physiology Sciences, Hainan medical college, Haikou, China
3Janta inter college Sohang-kushinagar, kushinagar, India
- *Corresponding Author:
- Yaoqing Chen
Jian Fu
Department of Pharmacology
Hainan medical college
Haikou China
E-mail: katotds@sina.com
- Yang Wang
Laboratory of Extreme Environment Sports, Medicine and Physiology Sciences
Hai Nan Medical College, Hai Nan Island, China
Tel: +0086-13807694497
E-mail: katotds@sina.com
Accepted date: December 13, 2018
Abstract
The hepatocellular carcinoma represent significant drug resistance to impediment effective treatment. The novel benzoxazole derivatives and the cytotoxicity effect toward hepatocellular carcinoma displayed critical regulatory process to cause cellular apoptosis. In this study, we investigated synthesized novel benzoxazole derivative dosage and time dependent bioaviability and morphology apoptosis in hepatoma cell (SMMC-7721). The derivative induced exposure of phosphatidylserine, DNA fragmentation and cleave of cysteine-aspartic acid protease expression in the execution of cell apoptosis were evaluated. The novel benzoxazole derivative dosage dependent SMMC-7721 cell membrane phosphatidylserine sensing to annexin a5 affinity were investigated by annexin a5 and propidium iodide dual staining flow cytometry. Cellular morphology of apoptosis were identified by Hoechst33258 stained adenine and thymine riches double-stranded DNA. The cytotoxicity induced DNA fragmentation in SMMC-7721 cell were analyzed by TdT-mediated dUTP nick-end labeling and agarose gel electrophoresis. Apoptosis relative cleaved cysteine-aspartic acid protease expression were quantified by westernblot. The results inducated that 2.5 μmol/L and 5 μmol/L was the optimum low dosage in genotoxicity of SMMC-7721 cell, however the significant decreasing of cellular viability in SMMC-7721 recognized in high dosages. The derivative induced phosphatidylserine sensing to annexin a5 affinity were increased 300% in the high dosage of 30 μmol/L. 24 hours incubation was the optimum time to obtain cellular morphological apoptosis and the significantly DNA ladder in optimum low dosage 5 μmol/L. DNA fragmentation were observed after 48 hours incubation in optimum low dosage. The A preliminaryprocaspase-3 and -9 down-regulated and the activated cleaved caspase-3, -9 up-regulated were recognized in the moderate dosage of 10 μmol/L, morevover become too significant in maximum dosage of 30 μmol/L. However, Poly (ADP-ribose) polymerase were preliminary significantly inhibited in the moderate dosage of 20 μmol/L. Conclusions: In vitro synthesized novel benzoxazole derived hepatocellular carcinoma SMMC-7721 cell morphological apoptosis were dosage-dependent that were observable in the lowest dosage of 2.5 μmol/L, however cellular phosphatidylserine exposure were presented in quadrupling of this lowest dosage. The derivative intervene manifested as DNA double strain chain breaks, DNA fragmentation and single-strand DNA breaks in low dosages, while decreasing the expression and cleaving of procaspase-3 and -9 in moderate dosages. Based on the above results, it can be concluded that synthesized novel benzoxazole derivative was the compound to introduce the genotoxicity in SMMC-7721 cell, further inducing apoptogenic effectiveness. This the important mechanism for this new structure benzoxazole toovercome the hepatic carcinoma drug resistance.
Keywords
Novel benzoxazole, hepatoma cell, genotoxicitic apoptosis, anti drugresistance of treatment.
Introduction
Several synthesized novel benzoxazole were demonstrated the potent role in cytotoxicity on carcinoma cells, especially in lung cancer cell [1], human hepatocellular cancer cell line (HePG- 2) and breast carcinoma cell line (MCF-7) [2,3]. Furthermore benzoxazole compound genotoxicity were identified to induce caspase-3 dependent apoptosis in HN-5 cancer cells [4], the inhibited RNA synthesis and interacted with the DNA molecule [5], and promising antitumor activity [6]. Benzoxazole anticancer mechanism were included inhibitory epidermal growth factor receptor activity. Caspase-3 and -9 protein level expression were cell cycle analysis showed pre G1 apoptosis and cell cycle arrest at G2/M phase. Up-regulation of Bax and down-regulation of Bcl-2 protein expression level confirmed apoptosis [7], effectively suppresses interleukin-6 activated STAT3 signaling pathway in immune cell [8]. Benzoxazole anticancer mechanisms were also reported in selectively inhibiting the activity of COX enzymes in breast carcinoma (MCF-7) and non-small cell lung cancer [9], inhibiting of macrophage migration inhibitory factor tautomerase active in DU-145 prostate cancer cells and pulmonary endothelial cells [10]. Hepatocellular carcinoma was a genetic disease with frequent somatic alterations in DNA. The anti-cancer treatment faced drug-resistance were the difficulties that are not easy to overcome. Therefore the genotoxicity will be a mentality of designing the structure of benzoxazole derivative. SMMC- 7721 was diseased type hepatoma cells line, analysis identified 29 recurrently amplified regions and 22 deleted regions with a high level of copy number changes of oncogenic drivers, harboring established oncogenes and tumor suppressors, including CCND1, MET, CDKN2A and CDKN2B, as well as many other genes not previously reported to be involved in liver carcinogenesis [11]. The combination treatment of benzoxazole analogues were reported to increase cytotoxicity enhanced antileukemic and apoptotic effects, increase in the expression levels of the pro-apoptotic (BAX, BAD and BIM) genes and hydrogen bonding interactions to important amino acids of BCR-ABL kinase receptor in K562R and K562S cells [12]. Benzoxazole derivatives as a negative regulator of apoptosis inhibiting micro RNAs, miR-2, miR-13 and miR-14 in activity at their target sites, which induce an increased caspase activity, and in turn influence the caspase dependent apoptotic pathway [13]. The novel caboxamycin derivatives were reported bearing distinct substitutions in the aryl ring have been generated and improved bioactive properties in comparison with caboxamycin [14]. These research development prompted the structure relative potential capacity of benzoxazole analogues of its genotoxicity in hepatoma cell, demonstrated the impressive apoptogenic effects besides the cytototicity. As an alternate treatment strategy for overcoming the drug resistance in treatment. we synthesized a novel benzoxazole derivative and investigated its genotoxicity induced apoptogenic effects in SMMC-7721 hepatoma cell.
Methods
Monitoring inhibition of hepatoma cells proliferation
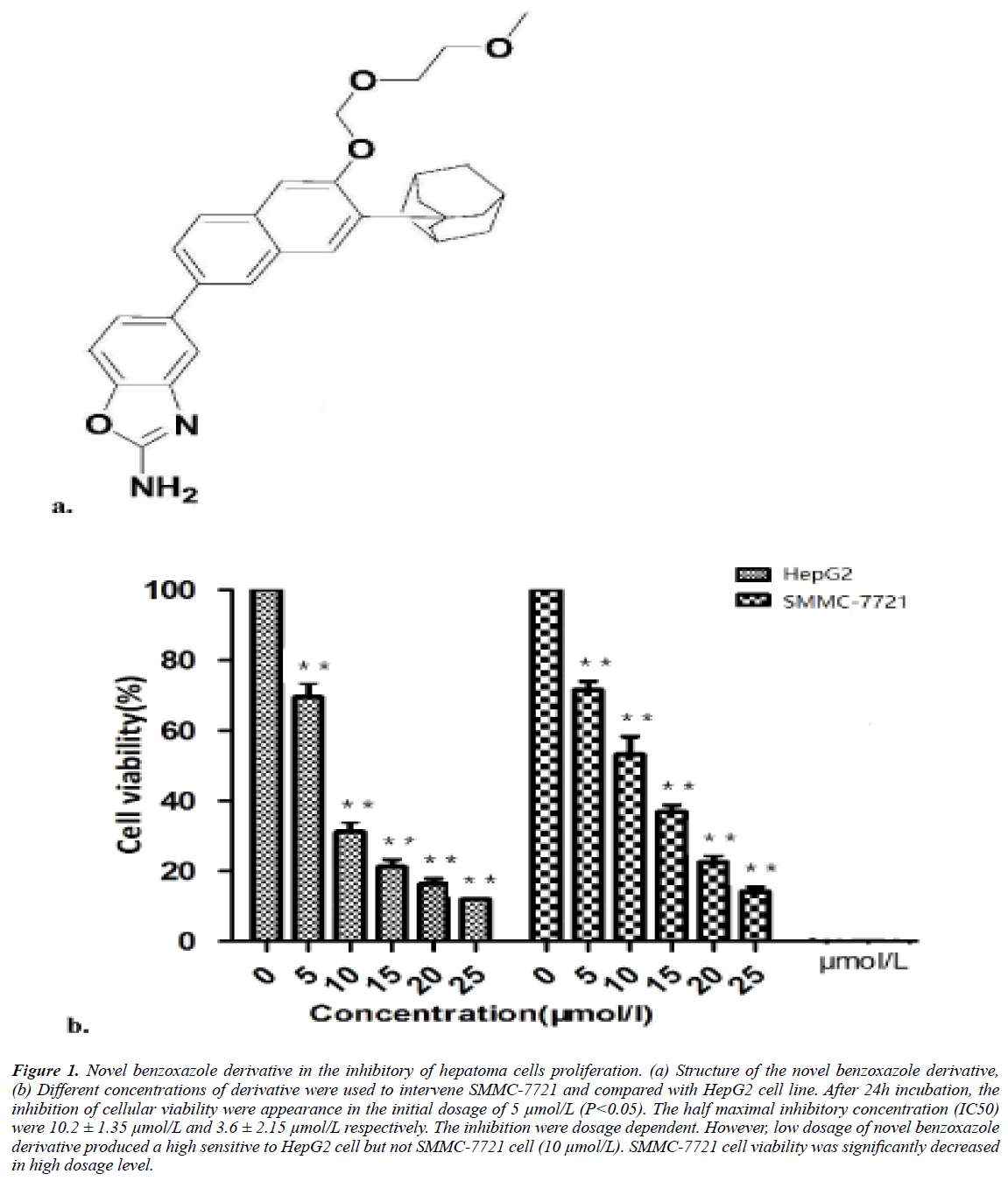
The novel synthesized benzoxazole were 5-{7-tricyclic [3.3.1.1 (3.7)] decanyl-6-(2-methoxy-ethoxymethoxy) -2-naphthyl}-2- amino benzoxazole. The structure shown in Figure 1a.
Figure 1: Novel benzoxazole derivative in the inhibitory of hepatoma cells proliferation. (a) Structure of the novel benzoxazole derivative, (b) Different concentrations of derivative were used to intervene SMMC-7721 and compared with HepG2 cell line. After 24h incubation, the inhibition of cellular viability were appearance in the initial dosage of 5 μmol/L (P<0.05). The half maximal inhibitory concentration (IC50) were 10.2 ± 1.35 μmol/L and 3.6 ± 2.15 μmol/L respectively. The inhibition were dosage dependent. However, low dosage of novel benzoxazole derivative produced a high sensitive to HepG2 cell but not SMMC-7721 cell (10 μmol/L). SMMC-7721 cell viability was significantly decreased in high dosage level.
Hepatoma cells line SMMC-7721 were from Shanghai institutes for biological sciences of Chinese academy of sciences. Cell line acquiring, storage and practicalities were under the guideline of Hainan medical college ethics committee. The cells were digested by trypsin, collected by centrifugation and counted. The cell concentration was adjusted to 2.5 × 104 cells/ml. The cells were then cultured in a cell suspension of 200 ml/pore of 5% CO2 and incubated in an incubator at 37ºC. After 24 hours of adherence, the cells were added with drugs of different gradient concentration. Each group was made into three holes and placed in culture. The cells were incubated in a carbon dioxide incubator for 4 hours with 10 ml cell counting kit-8 (CCK-8) solution. The absorbance values of the experimental group and the control group were measured on the enzyme label, and the detection wavelength was 450 nm. According to the formula, the inhibitory rate of cell proliferation was as follows: the inhibitory rate of cell proliferation (%)=(average OD of control group-average OD of experimental group)/average OD of control group *100%, and the half inhibitory concentration IC50 was calculated.
Hepatoma cells phosphatidylserine exposure and fractional DNA content
In vitro novel benzoxazole evoked hepatoma cells (SMMC- 7721) membrane phosphatidylserine sensing to annexin a5 affinity were investigated by annexin a5 and PI dual staining flow cytometry [15].
The cells were inoculated into 6-well plates with 1*107 cells per hole for 24 hours in an incubator with 37ºC, 5% CO2. After removing medium, the cells were treated with derivative-medium with final concentration of 0, 10, 20 and 30 μM, respectively. After 24 hours of derivative-medium incubation, the cells were digested with trypsin and centrifuged for 5 minutes at relative centrifugal force (RCF) 90 g to collecting the cells. Rings cells with PBS and then added 500 μL of binding buffer. In the final step, 5 μL of annexin V-FITC and 5 μL of PI working fluid was added. Mixture the solutions for 15 min at room temperature and flow cytometry analysis within 1 hour.
Cellular morphological apoptosi
The cellular morphology of apoptosis were identified by Hoechst33258 DNA staining. Tumor cells in good growth condition were used in the experiment. The aseptically treated cover slides were placed in the six-well plate, and the cells were inoculated into the six-well plate containing cover slides. The cells were 1*106/well, and then adhered to the plate for 24 hours. After treatment of cancer cells with different concentrations of drugs, the drug-containing medium was removed and 500 ml solid was added into each pore. Absorb the fixing fluid and wash it twice with PBS for 5 minutes each time. Absorb the liquid, add 500 ml Hoechst 33258 dye solution, place it in shaker, and dye for 5 minutes. Absorb the Hoechst 33258 dye solution and place it in a shaker. Wash it three times with PBS for 5 minutes each time. Absorbing the liquid, dropping anti-fluorescence quenching sealing solution on the slide, then covering the cover glass with cells on the slide, so that the cells contact with the sealing solution. The excitation wavelength was 350 nm by fluorescence microscopy. Fixed liquid for 20 minutes, and can stay overnight at 4ºC.
Apoptotic DNA fragmentation assay
DNA fragmentation in apoptosis cell were analyzed by agarose gel electrophoresis. Wash the sample slices three times with PBS and avoid light. The excitation wavelength was 450-500 nm by fluorescence microscopy. Add 50 ml POD-conjugated Anti- FITC working fluid to each sample, place it in a temperature box, avoid light, and react at 37ºC for 30 minutes. The samples were washed three times with PBS, and 50 ml DAB solution was added to each sample. The colour reaction lasted for 5 minutes. Then wash with PBS to terminate the reaction. Haematoxylin staining solution was added to the sample for 5 minutes. The samples were washed with distilled water, and then differentiated in 1% methanol hydrochloride solution for 5 seconds. The samples were washed with distilled water. Different concentration of ethanol was used for dehydration. Dimethylbenzene was soaked, dried, neutral gum was added and sealed. Photographs were observed under a microscope.
Terminal deoxynucleotidyl transferase dUTP nick end labeling assay
DNA fragmentation in apoptosis cell were analyzed by TdTmediated dUTP nick-end labeling [16]. Sample slices were soaked in 1% Triton X-100 permeate for 5 minutes. PBS was washed three times for 5 minutes each time. Samples were placed in 3% H2O2 sealing solution and sealed at room temperature for 10 minutes. Wash with PBS three times, 5 min each time. One group was treated with DNaseI as a positive control of apoptosis and incubated at 37ºC for 30 minutes. A group of TUNEL reaction liquids without TdT enzyme were used as negative control. They were placed in a temperature box, protected from light, and reacted at 37ºC for 1 hour. The samples were washed three times with PBS and labeled with Streptavidin- FITC working fluid of 50 ml. The samples were placed in a temperature box to avoid light and reacted at 37ºC for 30 min. Wash the sample slices three times with PBS and avoid light. The excitation wavelength was 450-500 nm by fluorescence microscopy. Add 50 ml POD-conjugated Anti-FITC working fluid to each sample, place it in a temperature box, avoid light, and react at 37ºC for 30 min. The samples were washed three times with PBS, and 50 μml DAB solution was added to each sample. The colour reaction lasted for 5 min. Then wash with PBS to terminate the reaction. Haematoxylin staining solution was added to the sample for 5 min. The samples were washed with distilled water, and then differentiated in 1% methanol hydrochloride solution for 5 seconds. The samples were washed with distilled water. Different concentration of ethanol was used for dehydration. Dimethylbenzene was soaked, dried, neutral gum was added and sealed. Photographs were observed under a microscope.
Cleaved caspase-3, -9 and poly (ADP-ribose) polymerase assay
The apoptosis relative caspase-3, -9 and poly (ADP-ribose) polymerase assay (PARP) expression were identified by westernblot. Extraction of total cell protein: After cells were treated with different concentrations of drugs, the treated cells were digested by trypsin, centrifuged, collected into centrifugal tube and placed on ice. Mix 1 mL RIPA lysate and 10 mu L PMSF to form cell lysate. Absorbing 200 ml of the above lysate and adding it to the centrifugal tube containing cell precipitation, blow it evenly, crack on ice for 3 hours, and take out several shocks every 30 minutes. Then centrifuge the supernatant at 4ºC for 15 min at 13000 r/min and then carefully transfer the supernatant protein solution to the new centrifugal tube. Store at -20ºC. Quantitative determination of protein concentration: BSA standard protein solution with final concentration of 0.5 mg/ml was prepared according to the instructions of the kit. The BSA standard protein solution of 0, 1, 2, 4, 8, 12, 16, 20 ml was absorbed and added to 96-well plate. PBS was added to 20-well plate to make the concentration of standard product 0, 0.025, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5 mg/ml respectively. Each group was made into three compound holes. Add 20 ml sample to 96-well plate, or dilute the sample to a certain multiple, add 3 multiple holes to each sample. According to the number of samples, BCA working liquid was prepared with 50:1 ratio of BCA reagent A and reagent B, and used within 24 hours. 200 ml BCA working solution was absorbed and added into each pore. The absorbency was determined by enzyme labeling at 37ºC for 30 min. The wavelength was 562 nm. The protein standard curve was drawn according to the optical density (OD) value and concentration of the standard protein solution, and the concentration was calculated from the OD value of the tested sample. The concentration of the sample to be measured is equal to the dilution multiple * the concentration of the diluted sample.
Statistical analysis
The results in the experiment were statistic as mean ± SEM. The differences between dosages were analyzed using unpaired t-test by Statistica (ver. 7, StatSoft).
Results
Novel benzoxazole inhibition of hepatoma cells proliferation
SMMC-7721 cell viability was inhibited by novel benzoxazole derivative in 5 μmol/L (P<0.05). The average of half maximal inhibitory concentration (IC50) was 10.2 ± 1.3 μmol/L, however in a parallel experiment, the IC50 of HepG2 cells were 6.8 μmol/L. However, SMMC-7721 cell viability was significantly decreased in high dosage level but not in the range of low dosages. In the dosage of 10 μmol/L and 15 μmol/L, the derivative were more effectiveness to HepG2 cell.
Cell phosphatidylserine exposure and fractional DNA content
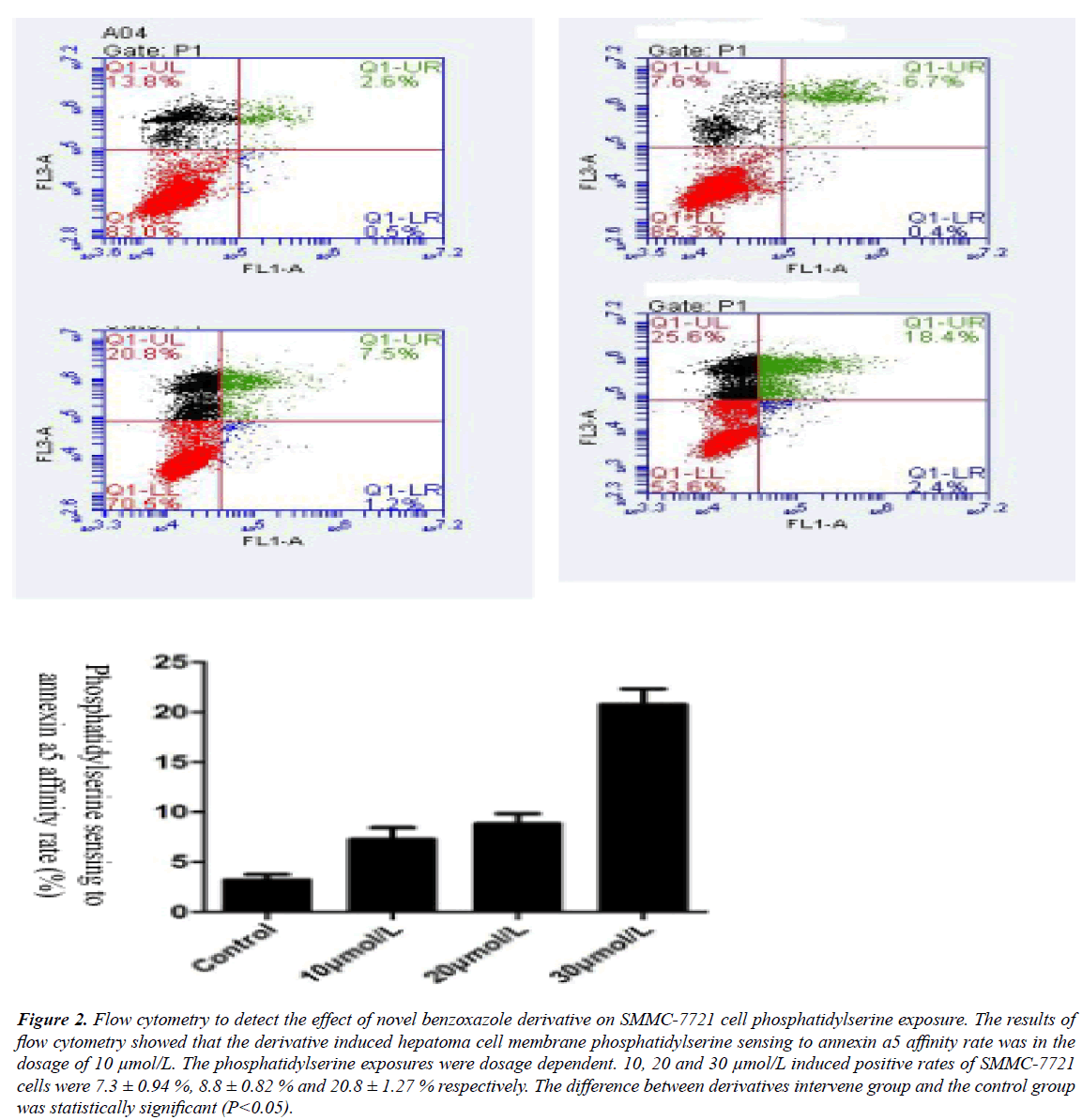
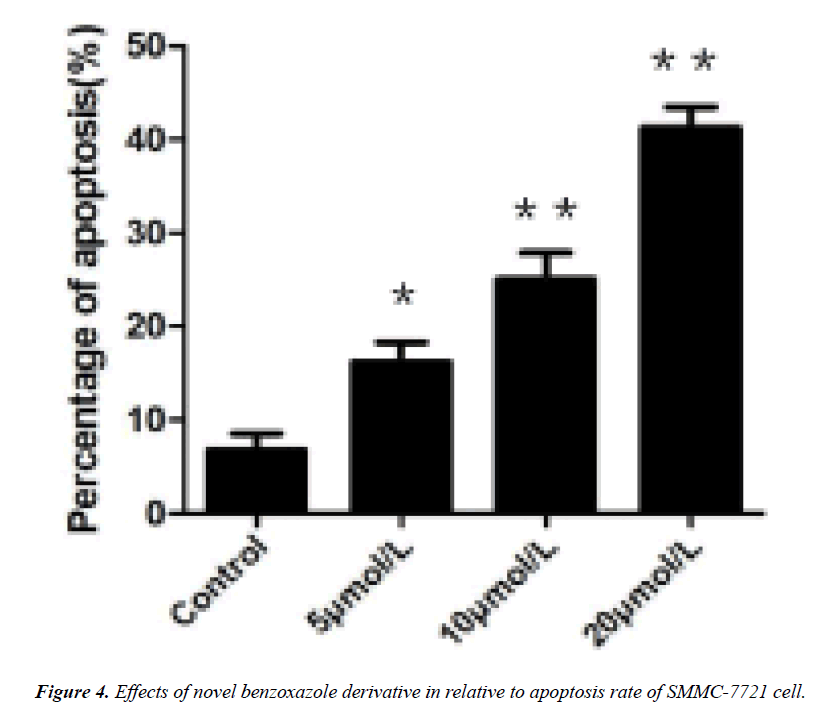
The results of flow cytometry showed that novel benzoxazole derivative could induce apoptosis of SMMC-7721 cells, and the higher the concentration of the compound, the higher the apoptotic rate. When the concentration of benzoxazole compound was 10, 20 and 30 μmol/L, the apoptotic rates of SMMC-7721 cells were (7.3+0.94)%, (8.8+0.82)% and (20.8+1.27)% respectively. Therefore, the benzoxazole compound can induce apoptosis of SMMC-7721 cells in a concentration-dependent manner. The higher the concentration of benzoxazole compound, the lower the proportion of nonapoptotic cells and the higher the proportion of apoptotic cells. The difference between the experimental group and the control group was statistically significant (P<0.05).
Novel benzoxazole induced Apoptotic DNA fragmentation
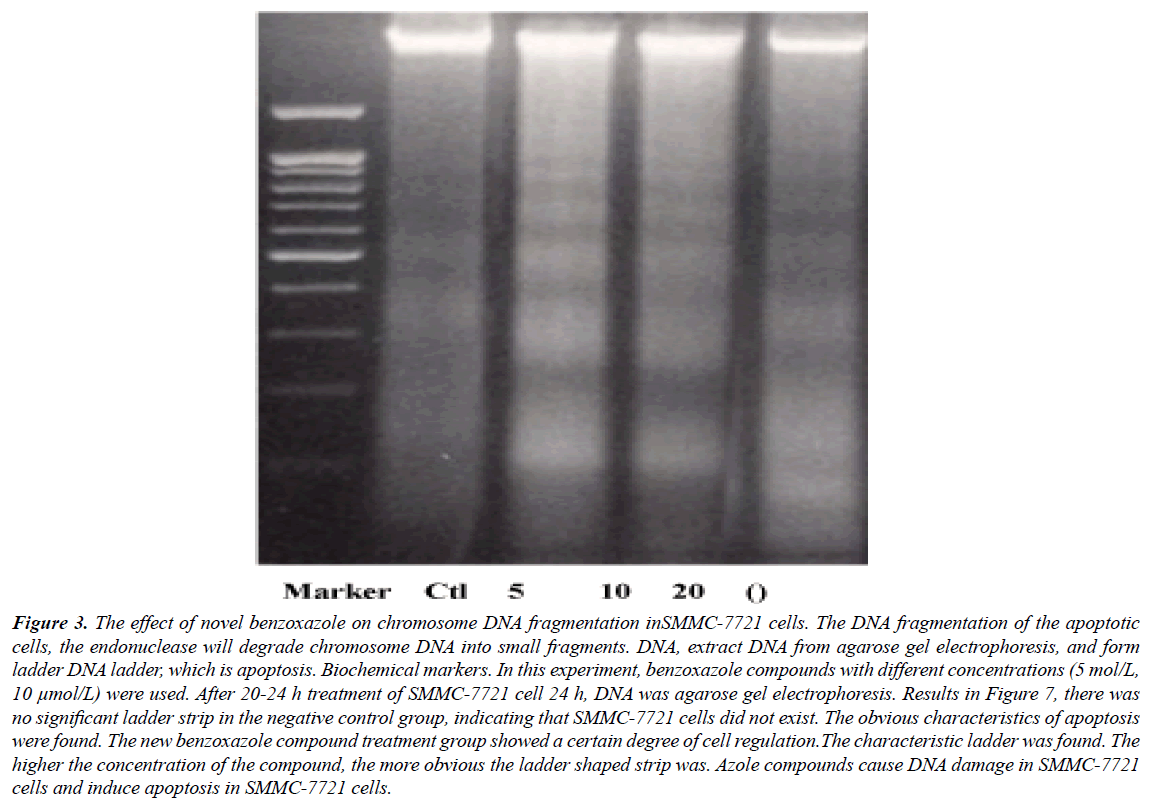
DNA agarose gel electrophoresis showed that the DNA fragmentation were observable in 5 μmol/L of derivative intervene and significantly in 20 μmol/L (Figures 1-3). TUNEL experiments showed that DNA 3'-hydroxyl termini of double strand breaks were increased in initial dosage of 5 μmol/L and significantly in 10 μmol/L and 20 μmol/L. This was contributed with cellular apoptosis rate within 24 hours (Figure 4).
Figure 2: Flow cytometry to detect the effect of novel benzoxazole derivative on SMMC-7721 cell phosphatidylserine exposure. The results of flow cytometry showed that the derivative induced hepatoma cell membrane phosphatidylserine sensing to annexin a5 affinity rate was in the dosage of 10 μmol/L. The phosphatidylserine exposures were dosage dependent. 10, 20 and 30 μmol/L induced positive rates of SMMC-7721 cells were 7.3 ± 0.94 %, 8.8 ± 0.82 % and 20.8 ± 1.27 % respectively. The difference between derivatives intervene group and the control group was statistically significant (P<0.05).
Figure 3: The effect of novel benzoxazole on chromosome DNA fragmentation inSMMC-7721 cells. The DNA fragmentation of the apoptotic cells, the endonuclease will degrade chromosome DNA into small fragments. DNA, extract DNA from agarose gel electrophoresis, and form ladder DNA ladder, which is apoptosis. Biochemical markers. In this experiment, benzoxazole compounds with different concentrations (5 mol/L, 10 μmol/L) were used. After 20-24 h treatment of SMMC-7721 cell 24 h, DNA was agarose gel electrophoresis. Results in Figure 7, there was no significant ladder strip in the negative control group, indicating that SMMC-7721 cells did not exist. The obvious characteristics of apoptosis were found. The new benzoxazole compound treatment group showed a certain degree of cell regulation.The characteristic ladder was found. The higher the concentration of the compound, the more obvious the ladder shaped strip was. Azole compounds cause DNA damage in SMMC-7721 cells and induce apoptosis in SMMC-7721 cells.
Cellular morphological apoptosis
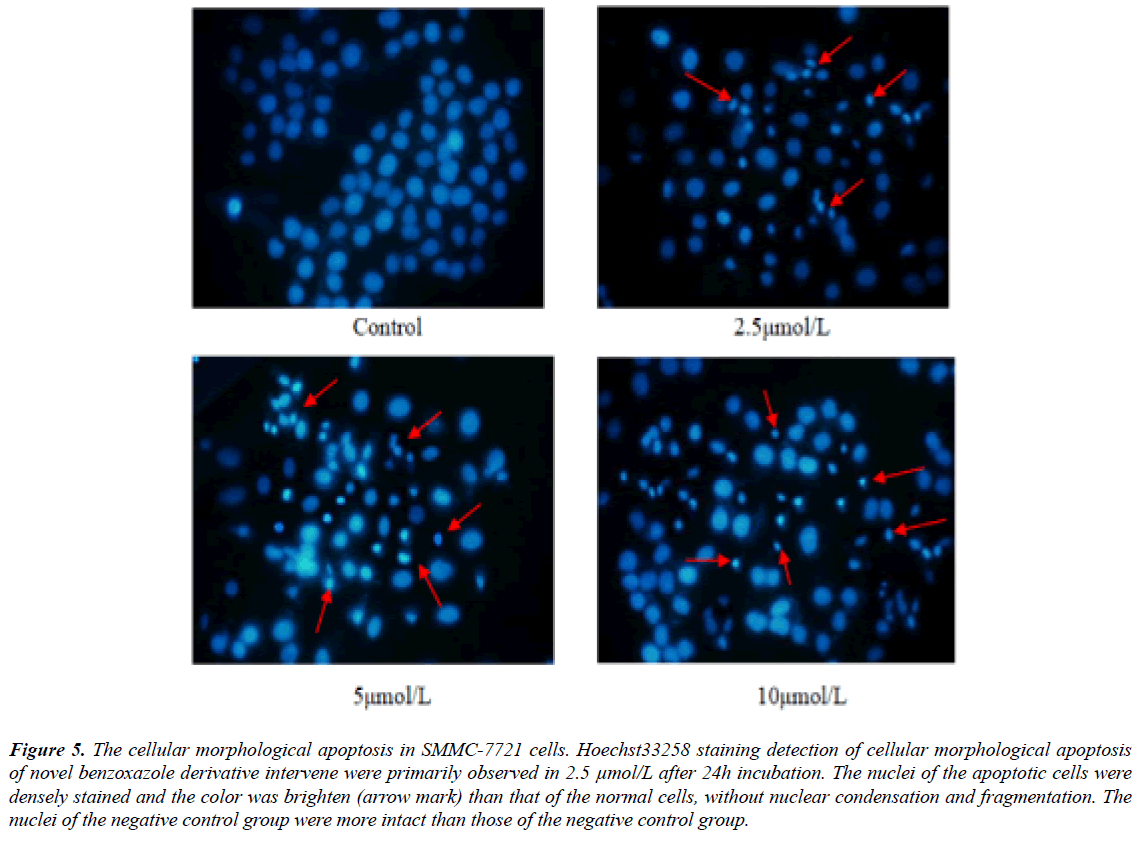
Hoechst33258 staining showed that cellular morphological apoptosis were from 24 hours after novel benzoxazole derivative intervene. Fluorescence microscopy showed that the nuclei of the apoptotic cells were densely stained and the color was brighten than that of the normal cells (Figure 5).
Figure 5: The cellular morphological apoptosis in SMMC-7721 cells. Hoechst33258 staining detection of cellular morphological apoptosis of novel benzoxazole derivative intervene were primarily observed in 2.5 μmol/L after 24h incubation. The nuclei of the apoptotic cells were densely stained and the color was brighten (arrow mark) than that of the normal cells, without nuclear condensation and fragmentation. The nuclei of the negative control group were more intact than those of the negative control group.
Apoptotic caspases and PARP assay
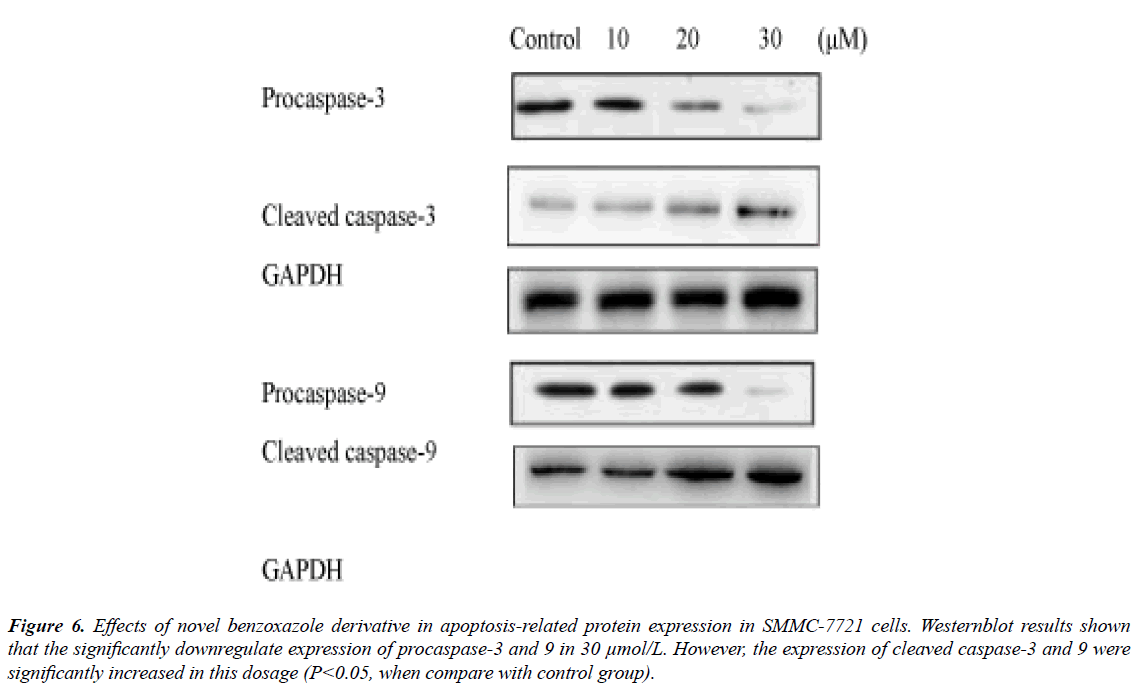
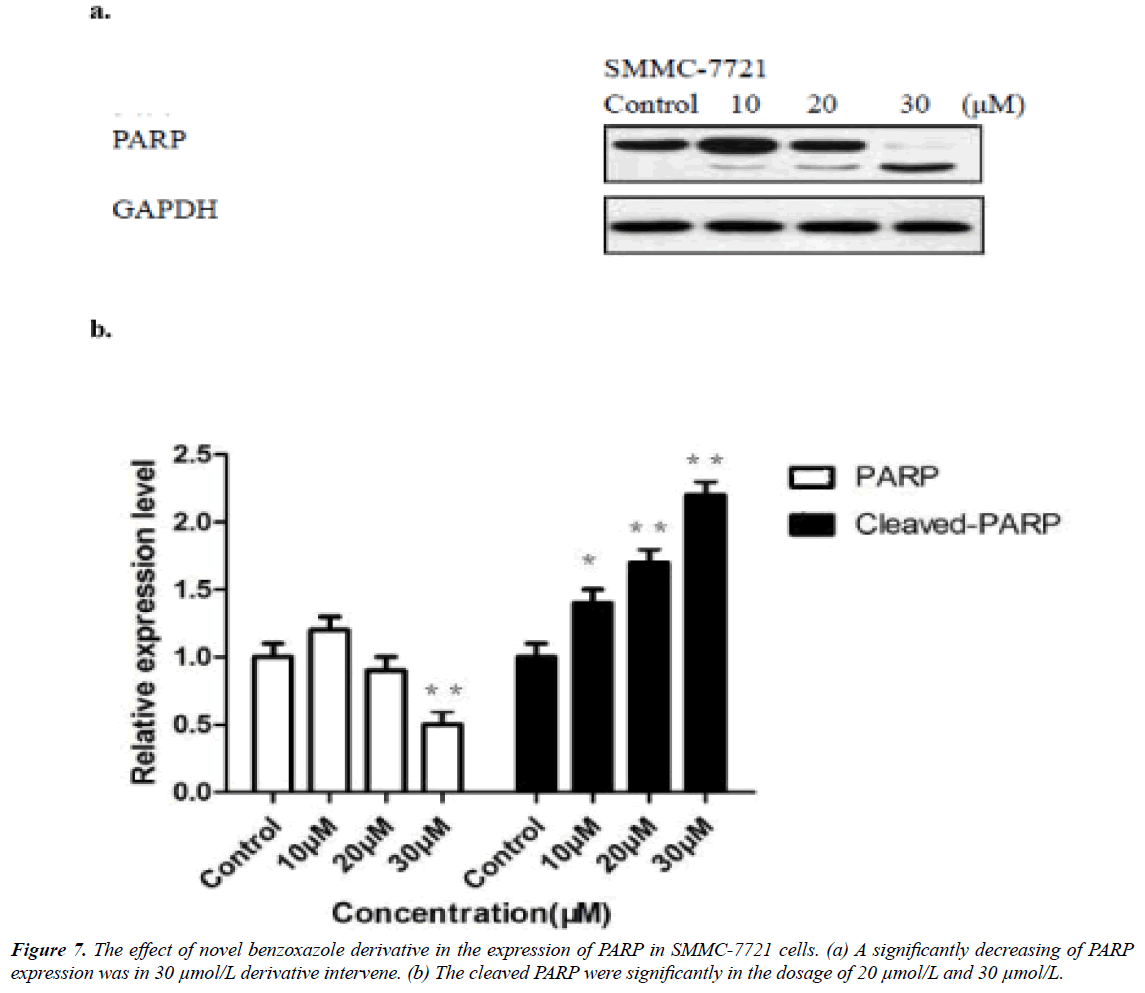
The novel benzoxazole derivative intervene induced related caspase-3 and -9 activations were from 10 μmol/L and significantly in 30 μmol/L (Figure 6), while the PARP significantly reduced in 30 μmol/L (Figure 6). The expression of Bcl-2 molecules in SMMC-7721 and HepG2 cells increased Bax expression, resulting in apoptosis (Figure 7).
Figure 6: Effects of novel benzoxazole derivative in apoptosis-related protein expression in SMMC-7721 cells. Westernblot results shown that the significantly downregulate expression of procaspase-3 and 9 in 30 μmol/L. However, the expression of cleaved caspase-3 and 9 were significantly increased in this dosage (P<0.05, when compare with control group).
Discussion
Benzoxazoles is a planar molecule with conjugated p electrons sextets in the cyclic systems. The lone pair of electrons on nitrogen, which is coplanar with heterocyclic ring and therefore not involved in delocalization, confers weakly basic properties. Targets containing the benzoxazole moiety, either isolated from plants or accessed by total synthesis have remarkable biological activities [17]. The heterocyclic nitrogen compound was first reported of its activation against leukemia [18]. After that, benzoxazole and pyridoimidazole derivatives were indicated to have the properties binding to target DNA sequence that were related to topoisomerase inhibitory properties [19]. The benzoxazole analogs was found binding with different amideand ester-linked side chains to duplex DNA in the presence of divalent metal cations that enhanced DNA binding of the ligands with the long side chains which exhibited the cytotoxicity of the lung and breast cancer cells [20]. The structurally unique bis (benzoxazole) natural product demonstrated the ability to bind a variety of di- and tri-valent metal ions, particularly Mg2+, and to form complexes with double-stranded DNA in the presence of Mg2+. The Hoechst33258 staining results recognized the quantitively decreased of DNA poly adenine and thymine sequence enrichment region in most low dosage (2.5 μmol/L, 24 hour incubation), therefore appeared apoptosis in low dosages. The analogue of natural benzoxazole isolated from Streptomyces in which the carbomethoxy-substituted benzoxazole ring was replaced with carbomethoxy-substituted benzimidazole ring was inactive against human cancer cell lines, however, a simplified analogue in which the carbomethoxy-substituted benzoxazole ring was replaced with a carbomethoxy group activated in against the cancer cell lines [21]. The new synthesized benzoxazole derivative avoided this naturally formed defect structure. It was reported pyrrolobenzodiazepine conjugates attached through different alkane or alkylamide spacers were promising DNA-binding ability and apoptosis caused G0/G1 phase arrest at sub-micromolar concentrations to induce the cancer cell apoptosis [22], although in this time the details of the new synthesized benzoxazole derivative mechanism is still not clear. The benzoxazole derivatives cytotoxicity were determined by its benzimidazole ring structure, The placement of methyl groups at the 5, 6 positions of the benzimidazole ring based on the lead structure 2-(benzimidazol-2-yl)-3-(5-nitrothiophen-2- yl) acrylonitrile lead to a 3-fold increase in overall cytotoxic activity, as wellreplacing the nitrothiophene for pyridine reduced cytotoxic activity as did replacing the nitro group for a methoxy group. Cytotoxic activity was only slightly reduced when the benzimidazole ring was replaced by a imidazo [4,5-b] pyridine or a benzthiazole ring but replacement by benzoxazole led to a substantial decrease in activity. Moving the acrylonitrile group from position 2 to position 1 of the benzimidazole ring also resulted in moderately active compounds. (Benzimidazol- 1-yl) acetamides showed only modest activity. These structure activity relationships found in the cytotoxicity studies are mirrored in the results of our novel benzoxazole derivative experiments.
The novel benzoxazole derivative in this study was 5-{7-tricyclic [3.3.1.1 (3.7)] decanyl-6-(2-methoxy-ethoxymethoxy)-2- naphthyl}-2-amino benzoxazole that included the unique acetoxy group on its side chain (Figure 1a). This unique structure indicated to primarily inhibit hepatocellular carcinoma line SMMC-7721 cell viability in the lowest dosage of 2.5 μmol/L by damage the DNA double strain (Figure 5). Nevertheless, fourfold increasing of dosage (10 μmol/L) of the derivative dosage result in phosphatidylserine exposure, and twelvefold dosage arriving exposure to maximum (Figure 2). This dosage related bioviability were suggested based on SMMC-7721 cell biological characteristics that originated from female hepatocellular carcinoma. The several cancer-related stresses and inhibiting apoptosis at multiple points in the apoptotic signal pathway were reported in SMMC-7721 cell that have its own pathways to contribute to the tumorigenesis, tumor growth and metastasis of liver cancer [23]. Several previous reports represented that the unique benzoxazole derivative structure induced carcinoma cell viability were relative to the cellular types, and further relative to the effectiveness of DNA fragmentation [24-26] to cleavage of the genomic DNA into oligonucleosomal fragments represented by multiples of 180- 200 bp. Visualizing these fragments can aid in characterizing hepatocellular carcinoma cells apoptotic event. SMMC-7721 cell were multi-drug resistance, such as significantly increasing the tumor volume in bevacizumab treated SMMC-7721 xenograft model that was reported by altogen labs [27]. In our experiment the 10 μmol/L of the derivative induced 20% apoptosis of the SMMC-7721 cell, however this dosage induced twofold high apoptosis in liver hepatocellular carcinoma HepG2 line (the data was not shown in this paper). Moreover, the novel benzoxazole derivative induced a significantly cleavage of genomic DNA and DNA 3'-hydroxyl termini of double strand breaks at low dosage of 5 μmol/L (Figure 3) which represented that this type of novel benzoxazole derivative were more target on DNA fragmentation. (Figure 4). The results furthermore indicated the derivative induced inhibition of hepatocellular carcinoma cells loss of viability were the apoptosis but not necrotic cell.
The new synthesized benzoxazole derivative also showed pro-apoptotic effect through activation of both intrinsic and extrinsic initiator caspases in female type SMMC-7721 cell. The caspases (cysteine-dependent aspartate-directed proteases) play essential roles in apoptosis which based on the cysteine in its active site, nucleophilically attacks and cleaves the target protein after residue of aspartic acid. Caspases mediated Fas receptor transduces apoptotic death signaling [28], while effectively inhibited caspase prevented CD95-mediated apoptosis and the potentiated TNF-induced necrosis 26) [29]. The caspase-3 significantly increased cyclic AMP (cAMP) response element-binding protein (CREB) to be significantly decreased in immortal epithelial-like tumorigenic cells HuH7 cells apoptosis [30]. The decreasing of caspase-3 activity were proved to be apoptosis of human hepatocellular carcinomas [31]. However, caspase-3 and -9 exists as the inactive form of zymogen in the cell, and were cleaved and activated by upstream caspases, then execute apoptosis by cleaving targeted cellular proteins. Caspase-3 cleaves multiple substrates including Poly (ADP-ribose) polymerase (PARP) that was activated in immediate cellular response to chemical induced single-strand DNA breaks. PARP binds to DNA and begins the synthesis of a polymeric adenosine diphosphate ribose chain which acts as the signal for the other DNA-repairing enzymes. Our results suggested that novel benzoxazole derivative active conjugates induced an increase in the expression of key apoptotic genes that are involved in the intrinsic pathway of caspase-3 and -9. The significant over-expression of cleaved caspase-3 and -9 protein level in treated samples was observed. Westernblot quantity analysis indicated that the low dosages significantly increased procaspase-3 and -9 was (3.75 μmol/L), as well as cleaved caspase-3 and -9. Moreover high dosage induced a significantly reduction of cell prototyping PARP (in 30 μmol/L). All these results indicated that novel benzoxazole derivative was capable in gene regulating level in the apoptotic pathway in hepatoma cells.
In conclusion, we suggested that the novel benzoxazole derivatives interaction with key apoptotic genes to activate the transcription of caspase-3 and -9 and its cell prototyping PARP expression. This further promote the apoptosis pathways in hepatoma cellsto improve the mechanisms in overcome the drug resistance.
Acknowledgements
This study is partly supported by Hainan province key R&D projects (ZDYF2017121).
Disclosure Statement
The authors declare that they have no competing interests.
References
- Al-Harthy T, Zoghaib WM, Pflüger M, et al. Design, Synthesis, and Cytotoxicity of 5-Fluoro-2-methyl-6-(4-aryl-piperazin-1-yl) Benzoxazoles. Molecules 2016; 21:E1290.
- Elhady HA, El-Sayed R, Al-Nathali HS. Design, synthesis and evaluation of anticancer activity of novel 2-thioxoimidazolidin-4-one derivatives bearing pyrazole, triazole and benzoxazole moieties. Chem Cent J 2018; 12:51.
- Sun M, Zhang X, Hao H, et al. Nocarbenzoxazoles A-G, Benzoxazoles Produced by Halophilic Nocardiopsis lucentensis DSM 44048. J Nat Prod 2015; 78:2123-7.
- Yaghmaei S, Ghalayani P, Salami S, et al. Hybrid Benzoxazole-Coumarin Compounds Induce Death Receptor-Mediated Switchable Apoptotic and Necroptotic Cell Death on HN-5 Head and Neck Cancer Cell Line. Anticancer Agents Med Chem 2017; 17:608-14.
- Hall IH, Peaty NJ, Henry JR, et al. Investigations on the mechanism of action of the novel antitumor agents 2-benzothiazolyl, 2-benzoxazolyl, and 2-benzimidazolyl hydrazones derived from 2-acetylpyridine. Arch Pharm (Weinheim) 1999; 332:115-23.
- Zahran MA, El-Essawy FA, Yassin SM, et al. Rapid and efficient synthesis of 4-substituted pyrazol-5-one under microwave irradiation in solvent-free conditions. Arch Pharm (Weinheim) 2007; 340:591-8.
- Labib MB, Philoppes JN, Lamie PF, et al. Azole-hydrazone derivatives: Design, synthesis, in vitro biological evaluation, dual EGFR/HER2 inhibitory activity, cell cycle analysis and molecular docking study as anticancer agents. Bioorg Chem 2018; 76:67-80.
- Kim D, Won HY, Hwang ES, et al. Synthesis of benzoxazole derivatives as interleukin-6 antagonists. Bioorg Med Chem 2017; 25:3127-34.
- Abdelgawad MA, Bakr RB, Omar HA. Design, synthesis and biological evaluation of some novel benzothiazole/benzoxazole and/or benzimidazole derivatives incorporating a pyrazole scaffold as antiproliferative agents. Bioorg Chem 2017; 74:82-90.
- Le Hiress M, Akagah B, Bernadat G, et al. Design, Synthesis, and Biological Activity of New N-(Phenylmethyl)-benzoxazol-2-thiones as Macrophage Migration Inhibitory Factor (MIF) Antagonists: Efficacies in Experimental Pulmonary Hypertension. J Med Chem 2018; 61:2725-36.
- Wang K, Lim HY, Shi S, et al. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology 2013; 58:706-17.
- Ozkan T, Hekmatshoar Y, Ertan-Bolelli T, et al. Determination of the apoptotic effect and molecular docking of benzamide derivative XT5 in K562 cells. Anticancer Agents Med Chem 2017.
- Mondal T, Lavanya AVS, Mallick A, et al. Novel Triazole linked 2-phenyl benzoxazole derivatives induce apoptosis by inhibiting miR-2, miR-13 and miR-14 function in Drosophila melanogaster. Apoptosis 2017; 22:786-99.
- Losada AA, Méndez C, Salas JA, et al. Exploring the biocombinatorial potential of benzoxazoles: generation of novel caboxamycin derivatives. Microb Cell Fact 2017; 16:93.
- Darzynkiewicz Z, Bruno S, Del Bino G, et al. Features of apoptotic cells measured by flow cytometry. Cytometry 1994; 13:795.
- Lebon C, Rodriguez GV, Zaoui IE, et al. On the use of an appropriate TdT-mediated dUTP-biotin nick end labeling assay to identify apoptotic cells. Anal Biochem 2015; 480:37-41.
- Dhani R. Benzoxazole: the molecule of diverse pharmacological activities. Pharma research library.
- Bahner CT, Rives LM, McGaha SW, et al. Di- and tri-methoxystyryl derivatives of heterocyclic nitrogen compounds.Arzneimittelforschung 1981; 31:404-6.
- Bailly C, Colson P, Houssier C, et al. Mode of DNA binding of bis-benzimidazoles and related structures studied by electric linear dichroism. J Biomol Struct Dyn 1994; 12:173-81.
- Mazzitelli CL, Rodriguez M, Kerwin SM, et al. Evaluation of metal-mediated DNA binding of benzoxazole ligands by electrospray ionization mass spectrometry. J Am Soc Mass Spectrom 2008; 19:209-18.
- Kumar D, Jacob MR, Reynolds MB, et al. Synthesis and evaluation of anticancer benzoxazoles and benzimidazoles related to UK-1.Bioorg Med Chem 2002; 10:3997-4004.
- Kamal A, Reddy KS, Khan MN, et al. Synthesis, DNA-binding ability and anticancer activity of benzothiazole/benzoxazole-pyrrolo[2,1-c][1,4]benzodiazepine conjugates. Bioorg Med Chem 2010; 18:4747-61.
- Feng Y, Tian ZM, Wan MX, et al. Protein profile of human hepatocarcinoma cell line SMMC-7721: identification and functional analysis. World J Gastroenterol 2007; 13:2608-14.
- Özdemir F, Akalın G, Şen M, et al. HepG2 cell:Towards novel anti-tumor strategies for hepatic cancer: ɛ-viniferin in combination with vincristine displays pharmacodynamic synergy at lower doses in HepG2 cells. OMICS 2014; 18:324-34.
- Takashina M, Inoue S, Tomihara K, et al. Different effect of resveratrol to induction of apoptosis depending on the type of human cancer cells. Int J Oncol 2017; 50:787-97.
- Gozzi GJ, Pires Ado R1, Valdameri G, et al. Selective Cytotoxicity of 1,3,4-Thiadiazolium Mesoionic Derivatives on Hepatocarcinoma Cells (HepG2). PLoS One 2015; 10:e0130046.
- http://altogenlabs.com/xenograft-models/liver-cancer-xenograft/smmc-7721-xenograft-model
- Suzuki A, Tsutomi Y, Akahane K, et al. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene 1998; 17:931-9.
- Los M, Mozoluk M, Ferrari D, et al. Activation and caspase-mediated inhibition of PARP: a molecular switch between fibroblast necrosis and apoptosis in death receptor signaling. Mol Biol Cell 2002; 13:978-88.
- Hara M, Takeba Y, Iiri T, et al. Vasoactive intestinal peptide increases apoptosis of hepatocellular carcinoma by inhibiting the cAMP/Bcl-xL pathway. Cancer Sci 2018; 3.
- Yamanaka T, Shiraki K, Sugimoto K, et al. Chemotherapeutic agents augment TRAIL-induced apoptosis in human hepatocellular carcinoma cell lines. Hepatology 2000; 32:482-90.