Research Article - Biomedical Research (2017) Volume 28, Issue 8
Knockdown of ET-1 gene can inhibit the proliferation, invasion of human prostate cancer cell
Jie Liu and Xiuheng Liu*Department of Urology, Renmin Hospital of Wuhan University, Wuhan 430060, Hubei Province, PR China
- *Corresponding Author:
- Dr. Xiuheng Liu
Department of Urology
Renmin Hospital of Wuhan University
Hubei Province, PR China
Accepted on September 29, 2016
Abstract
Prostate cancer is the second most frequently diagnosed cancer and the sixth most commonly fatal cancer in men worldwide. Endothelin-1 (ET-1), produced by the prostate epithelia, likely plays an important role in the progression of prostate cancer. ET-1 has been previously reported in prostate and other organs. However, ET-1 expression in Prostate cancer cell lines has not been described. Therefore, we have used Western Blot to demonstrate ET-1 expression in breast cancer cell lines (LNCap cell line, PC3 cell line and DU145 cell line). The results suggest the expression of ET-1 is significantly high expressed in PC3 cell line. Moreover, knockdown of ET-1 gene with siRNA can effectively repress the cell proliferation and the cell cycle was blocked in G1/G2 phase. Knockdown of ET-1 gene can affect cell invasion, which is confirmed by cell invasion and migration related genes using west blot (-catenin, Ecadherin, RhoC and RhoA). Characterization of the role of ET-1 in the development of tumors could lead to a better understanding of the changes occurring at the molecular level during the development and progression of primary human Prostate cancer.
Keywords
Prostate cancer, Western blot, Cell proliferation, Human, Cell cycle.
Introduction
Prostate cancer is characterized by low rates of cell proliferation, coupled with diminished rates of cell death [1]. This pattern has made prostate cancer to be one of the most resistant malignancies against cytotoxic chemotherapeutic agents. In 2016, prostate cancer is expected to be diagnosed in 180,890 men, and 26,120 will die of metastatic disease [2]. While the majority of localized prostate cancers can be controlled with surgery and/or radiation, metastatic disease remains a lethal disease with no curative options. Moreover, prostate cancer is a heterogeneous disease that can be highly lethal but also slow and indolent, as reflected by a 10-year estimated survival of 17% [2], which make a reflection of underlying genomic diversity. Large-scale cancer genome characterization projects studying glioblastoma, lung, colon, pancreas and breast cancers have provided critical new insights into the molecular classification of cancers and have the potential to identify new therapeutic targets [3-5]. However, prostate cancer presents special challenges for such large-scale multicenter genomics projects because of the relatively small tumor size and admixture with stroma that requires careful pathologist-guided dissection. A number of groups have reported analyses of transcriptomes and copy-number variations (CNVs) in prostate cancer, but rarely from the same samples and typically from modest numbers of tumors (50-100 samples) or with lower resolution platforms [6-8]. Numerous transcriptome studies have defined general prostate cancer signatures; however, unlike breast cancer [9,10], these analyses have not identified robust subtypes of prostate cancer with different prognoses [11,12]. The advent of affordable and efficient techniques for profiling tumors molecularly represents an unprecedented opportunity to better characterize the molecular factors that result in indolent and/or lethal disease and to tailor therapy accordingly. For now, many clinical trials are already underway to examine whether molecularly targeted therapies can improve outcomes [13].
The potent vasoconstrictor peptide endothelin-1 (ET-1) was first isolated from the culture media of porcine endothelial cells in 1988 [14]. It is one of a family of multifunctional peptides (ET-, 2 and 3) that are closely related to the sarafotoxins derived from the venom of the burrowing asp. ET-, like its relatives ET-2 and ET-3, is a 21-amino-acid peptide characterized by a single alpha-helix and two disulphide bridges. The three endogenous endothelins are encoded by distinct genes and are regulated at the level of mRNA transcription. The primary translation product of the ET-1 gene is the 212-amino-acid prepro-ET-, which is cleaved by an endopeptidase to form the 38-amino-acid big-ET-1. The biologically active ET-1 is formed by endothelin-convertingenzyme (ECE) [15]. The half-life of ET-1 in circulation is seven minutes [16]. Of these isoforms, ET-1 has been the most extensively studied to date, and has been implicated in cancer. Two pathways have been described for clearance of endothelin: ET-B receptor-mediated uptake followed by lysosomal degradation [17,18] and catabolism by extracellular neutral endopeptidase (NEP) [19]. Endothelin production is stimulated by a variety of cytokines and growth factors, including IL-1β [20], TNF-α [21], TGF-β [22], platelet derived growth factor [23], vasopressin [24], prostate specific antigen [25] and low-density lipoprotein [26]. Local low oxygen tension and shear stress on vessels walls also increase ET-1 production [27,28]. Inhibitory factors include nitric oxide, prostacyclin, and atrial natriuretic hormone. ET-1 has been implicated in prostate cancer disease progression [29]. ET-1 expression occurs in almost every human prostate cancer tissue [30]. Moreover, patients with metastatic prostate cancer have elevated levels of plasma ET-1 compared with patients with organ-confined cancer as well as healthy individuals [31]. Activation of the endothelin receptor A (ETA) can lead to induction of a survival pathway, whereas activation of the endothelin receptor B (ETB) can result in clearance of circulating ET-1 as well as in stimulation of apoptosis. However, the response to the binding of either receptor remains cell type-dependent. In prostate cancer, the expression of the endothelin receptors, ETA and ETB, is altered compared to the pattern seen in normal prostatic tissues [30]. The ETB, predominant on benign prostatic epithelial cells, has a much lower expression on prostate cancer cells, owing, at least in part, to frequent hypermethylation of the ETB gene, EDNRB [32]. Increased ET-1 expression, coupled with the increased ETA expression that occurs with higher prostate tumor stage and grade, may produce a survival advantage for the prostate cancer cells. In studies of endothelial and stromal cell populations, ET-1 acting through the ETA inhibited apoptosis induced by a cytotoxic agent [33]. Given that endothelin receptor expression in prostate cancer favors the ETA and the compelling results from the atrasentan clinical trials, it is our hypothesis that ET-1 can act as a survival factor for prostate cancer. Therefore, we investigated the effects of ET-1 knockdown in PC-3 cell line in human prostate cancer cell line. Characterization of the role of ET-1 in the development of tumors could lead to a better understanding of the changes occurring at the molecular level during the development and progression of primary human prostate cancer.
Results
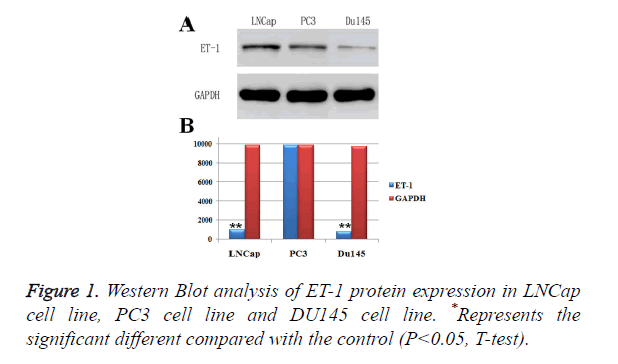
ET-1 was high expressed in PC3 cell lines
In order to investigate the conditions of ET-1 expressions in prostatic cancer cell lines, LNCap cell line, PC3 cell line and DU145 cell line were used to perform Western Blot analysis of ET-, and GAPDH was employed as a control (Figure 1A). As shown in Figure , the ET-1 protein was detectable in all prostatic cancer cell lines. However, the highest expression level of ET-1 protein appeared on PC3 cell line and LNCap cell line and the lowest expression level of ET-1 protein appeared on DU145 cell line, indicating that ET-1 protein in PC3 cell and LNCap cell was high expressed. Moreover, protein expressions of ET-1 were confirmed by gray value analysis, which was similar with the previous results in three different prostatic cancer cell lines (Figure 1B). However, the ET-1 protein expression in LNCap cell line was significantly differences compared with the GAPDH, which was contradictory with the previous analysis. In summary, the highest protein expression level of ET-1 appeared on the PC3 cell line.
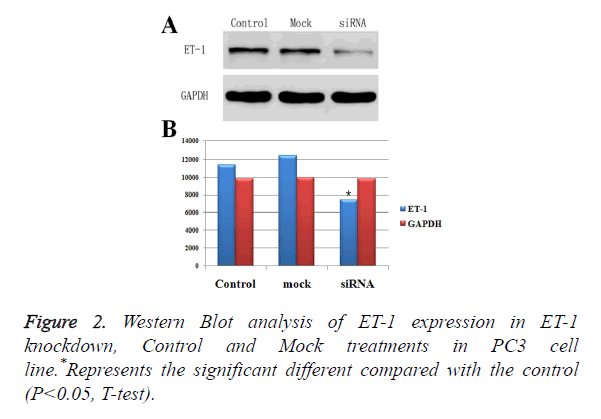
Effect of ET-1 knockdown in PC3 cells
To down regulate ET-1 expression in PC3 cells, we designed a siRNA against ET-1. The ET-1 siRNA was used to transfect PC3 cells, and Western Blotting analysis was performed to measure ET-1 expression. The results showed that ET-1 siRNA effectively decreased ET-1 expression (Figures 2A and 2B). Moreover, protein expressions of ET-1 in experimental group were significant difference compared with the Control group and Mock group, which was confirmed by gray value analysis (Figure 2B). Therefore, ET-1 siRNA were used in all subsequent experiments.
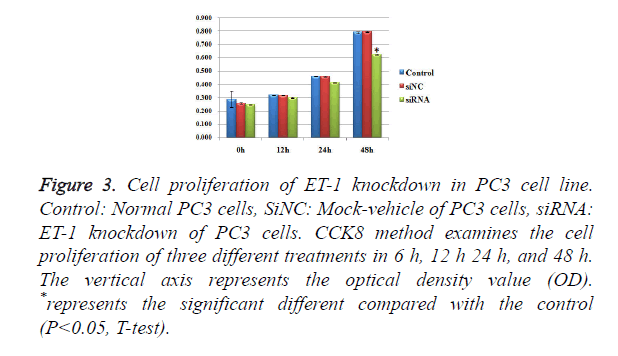
Influence of treatments with ET-1 knockdown on PC3 cell proliferation
To test whether ET-1 knockdown might affect PC3 cell proliferation, we introduced three treatments of Control, SiNC and SiRNA and observed the conditions of cell proliferation in 0h, 12 h, 24 h and 48 h (Figure 3), respectively. There were no significant differences of cell proliferation between the three different treatments in 0 h, 12 h and 24 h. For the group in 48 h, SiRNA treatments showed significant differences of cell proliferation compared with the control group and SiNC group. However, there was no significant difference between the SiNC treatments and the control group. The results mentioned above suggested that ET-1 knockdown treatments on PC3 cells can effectively repress the cell proliferation.
Figure 3. Cell proliferation of ET-1 knockdown in PC3 cell line. Control: Normal PC3 cells, SiNC: Mock-vehicle of PC3 cells, siRNA: ET-1 knockdown of PC3 cells. CCK8 method examines the cell proliferation of three different treatments in 6 h, 12 h 24 h, and 48 h. The vertical axis represents the optical density value (OD). *represents the significant different compared with the control (P<0.05, T-test).
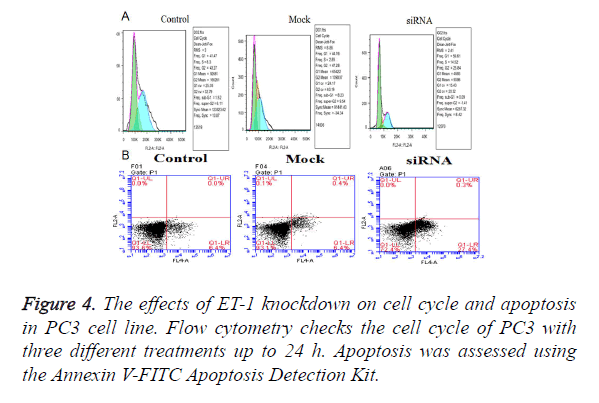
Effect of ET-1 knockdown on cell cycle regulation and apoptosis in PC3 cell
The effects of ET-1 knockdown on PC3 cell cycle progression and apoptosis were analyzed by flow cytometry. The Fluorescence Activated Cell Sorter (FACS) results indicated that ET-1 knockdown in PC3 cells induced a transient cell accumulation in G1 and G2 phase (Figure 4A), while ET-1 knockdown resulted in accumulation of cells in G1 phase coincident with a cell decrease in G1 phase up to 48 h of transfection (Figure 3), suggesting a block at G1/G2 check point. Cell proliferation of ET-1 knockdown in PC3 cells was significantly refrained compared with the Mock and Control groups up to 48 h. The results mentioned above suggested that ET-1 knockdown on PC3 cells can effectively repress the cell proliferation by preventing the cell cycling of G1/G2 phase. Moreover, ET-1 siRNA resulted in a significant increase in apoptosis compared with Control and Mock groups; ET-1 knockdown significantly promoted apoptosis (Figure 4B).
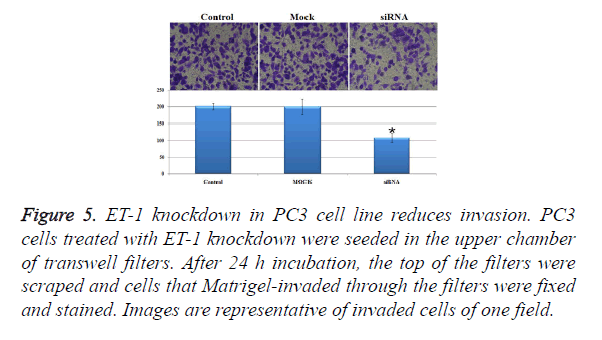
Influence of treatments with ET-1 knockdown on PC3 cell invasion
In order to investigate the invasion effect of ET-1 knockdown on PC3 cells, we used transwell chambers to assess the in vitro invasion potential of the tumor cells. In the cell invasion analysis, the number of invasion cells in siRNA treatment group was about half of the control group and the Mock group, and the number of invasion cell of siRNA treatment was significantly decreased when compared with the control group and the Mock group (Figure 5). The results mentioned above indicated that ET-1 knockdown treatment can effectively repress the cell invasion.
Figure 5. ET-1 knockdown in PC3 cell line reduces invasion. PC3 cells treated with ET-1 knockdown were seeded in the upper chamber of transwell filters. After 24 h incubation, the top of the filters were scraped and cells that Matrigel-invaded through the filters were fixed and stained. Images are representative of invaded cells of one field.
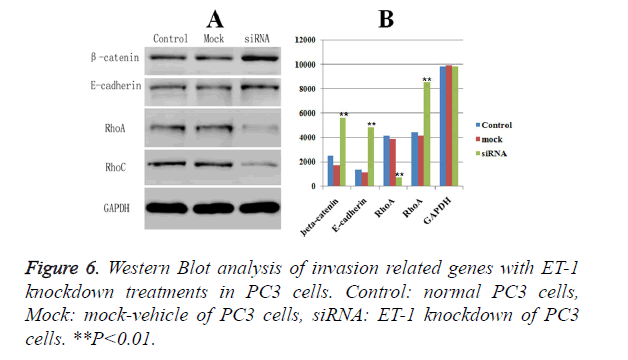
Western blot analysis of biomarkers related with invasion and migration of PC3 cell
The expressions of biomarkers related with invasion and migration, including -catenin, E-cadherin, RhoA and RhoC, were examined by Western Blot, respectively. GAPDH was served as the loading control. Western blot analysis showed that ET-1 knockdown treatment caused a significant down-regulated protein expression of the RhoA (Figure 6) compared with Control group and Mock group. Meanwhile, the protein expressions of the other three markers (β-catenin, E-cadherin and RhoC) were significant up-regulated in ET-1 knockdown treatment compared with the Control group and the Mock group. Moreover, there was no influence on the expressions of β-catenin, E-cadherin, RhoA and RhoC in the Control group and the Mock group. However, ET-1 knockdown treatment could lead to the significant down-regulation of the expression of RhoA and up-regulation of the expression of β-catenin, E-cadherin, RhoC.
Discussion
Endothelin-1 (ET-1), first isolated from porcine endothelial cells, is the most known potent endogenous vasoconstrictor, unlike its nearest competitor, angiotensin II (ET-1 is about 10 times more potent on a molar basis), ET-1 has prolonged effects [14]. The endothelin family contains several 21-amino acid members, all characterized by two defining disulphide bridges, [34] including ET-, ET-2 and ET-3, and the sarafotoxins (isolated from the venom of the Israeli burrowing asp Atractaspis engaddensis) [34,35]. These ligands bind to the ET receptors, ETA and ETB, with varying affinity [36]. The ETs are identical in all mammals and many higher vertebrates; the ET receptors are also very similar. Endothelin-1 is produced by many different cell types [23]. The pattern of ET-1 expression is unique among other neuroendocrine bioactive peptides, which have scattered, scant or undetectable expression in the normal prostate. Indeed, the concentrations of immunoreactive ET-1 are highest in seminal fluid, about 500 times greater than the concentration in circulation [37]. Endothelin-1 is expressed in prostatic tissues in vivo [38] and cultured in prostate cells [39]. The direct growth-promoting effects are modest, but ET-1 also synergized the proliferative effects of other peptide growth factors in certain prostate cancer cell lines [31]. Ligands for G-protein-coupled receptors, like ET-, can transactivate other, more classic, mitogenic pathways; ET-1 induces rapid tyrosine phosphorylation of EGFR in Rat-1 cells, in the absence of EGF itself [40]. ET-1 also elevates intracellular calcium in several prostate cancer cell lines [41]. Benign prostatic epithelium expresses the ETB receptor subtype predominantly [42], but in prostate cancer cells the ETB receptor is not expressed. Based on these data, blocking ET-1 action in prostate cancer should be through the ETA receptor, as the ETB receptor is frequently “silenced”. In order to gain understanding into the molecular events of ET-1 underlying the development of prostatic cancer, we analyzed the protein expressions of ET-1 in three different prostatic cancer cell lines (PC-3, DU145, LBCaP) to harvest the high expression cell line. Moreover, Effects of ET-1 knockdown in PC-3 cells were also performed. In the present study, Western Blot analysis clearly demonstrated that ET-1 knockdown in PC-3 cells can make the cell cycle distribution be retained in G1/G2 phase and cell proliferation was significantly refrained compared with the Mock and Control groups up to 48 h. Meanwhile, ET-1 siRNA resulted in a significant increase in apoptosis compared with Control and Mock groups; ET-1 knockdown significantly promoted apoptosis. In addition, the invasion analysis by Boyden chamber showed that ET-1 knockdown reduced cell invasiveness up to 24 h, which were confirmed by the western blot of biomarkers related with cell invasion. The protein expressions of biomarkers showed that ET-1 knockdown treatment caused a significant down-regulated protein expression of the RhoA compared with Control group and Mock group. Meanwhile, the protein expressions of the other three markers (beta-catenin, E-cadherin and RhoC) were significant up-regulated in ET-1 knockdown treatment compared with Control group and Mock group. Moreover, there was no influence on the expressions of beta-catenin, E-cadherin, RhoA and RhoC in the Control group and Mock group. However, ET-1 knockdown treatment could lead to the significantly down-regulation of the expression of RhoA and up-regulation of the expression of beta-catenin, E-cadherin, RhoC. In the present study, we investigated the regulation of ET-1 knockdown in PC-3 cell line, and the mechanisms underlying its regulation in human prostate cancer cell line. Characterization of the role of ET-1 in the development of tumors could lead to a better understanding of the changes occurring at the molecular level during the development and progression of primary human prostate cancer.
Method and Materials
Cell treatment
PC-3 cell line, DU145 cell line and LBCaP cell line were purchased from ATCC (Virginia, USA), and maintained in RPMI 1640 with 10% (v/v) FBS (Invitrogen, Carlsbad, CA). Cell lines were maintained in a humidified chamber at 5% CO2, at 37. Double-stranded siRNAs (dsRNA) targeting the ET-1 gene (NM_001168319) and complementary dsRNA were synthesized by Chemical Methods (ReiBo Biotech, China). The nucleotide sequence of the dsRNA for human ET-1 mRNA was 5’-ggg ucu aca uca acu auu a.dTdT-3’and 3’-dTdTccc aga ugu agu uga uaa u-5’. The nucleotide sequence of the control siRNA from a scramble sequence was 5’-uuc ucc gaa cgu guc acg u.dTdT-3’ and 3’-dTdT aag agg cuu gca cagugc a-5’. Cells were seeded at 5 × 105 cells per well in 6-well plates in DMEM containing 10% FBS without penicillin and streptomycin overnight. Transfection experiments were performed with OPTI-MEM serum-free medium and Lipofectamine 2000 reagent with a final siRNA concentration of 50 or 100 nM. The cells were collected for MTT assay and protein extraction after 24 h of siRNA transfection.
Cell proliferation assays
A cell proliferation assay was conducted with MTT kit (Sigma) according to the manufacturer’s instruction. For the colony formation assay, 500 cells were placed into each well of 6-well plate and maintained in media containing 10% FBS for 2 weeks. Colonies were fixed with methanol and stained with 0.1% crystal violet (Sigma) in PBS for 15 minutes. Colony formation was determined by counting the number of stained colonies in 3 randomly selected fields with an inverter microscope. Triplicate wells were measured in each treatment group.
Cell cycle analysis
Flow cytometric analyses were performed to define cell cycle distribution for transfected and not transfected cells. After 6, 12, 24 and 48 h from transfection, cells were harvested by trypsinization and fixed with 70% ethanol. Cells were stained for total DNA content with a solution containing 20 μg/ml propidium iodide. Cell cycle distribution was then analyzed with a FACS calibur flow cytometer (BD Biosciences).
Invasion assay
Cell invasion was analyzed by using Cultrex 24-well BME Cell Invasion Assay (Trevigen Inc., Gaithersburg, MD, USA) according to standard procedures. Briefly, 103 PC3-1 cells were seeded in 100 μl serum-free media into the upper wells previously coated with Matrigel basement extract, and 500 μl of media were added in the bottom wells. After 24 h of CO2 incubation at 37°C, the non-invasive cells on the upper surface were removed and the cells migrated to the lower surface were fixed in 500 μl of Cell Dissociation Solution/Calcein-AM, incubated al 37°C in CO2 incubator for 1 h and quantified by fluorimetric analysis (485 excitation, 520 nm emission).
Western blot analysis
Total cellular protein in three different treatments was isolated by the addition of 1% PMSF and RIPA lysis buffer (50 mM Tris-HCl (pH 7.4), 150 m MNaCl, 1% NP-40, 0.1% SDS). After boiled with SDS-PAGE sample buffer for 5 min, the samples were performed for sodium dodecylsulfate-polyacrylamide gel electrophoresis. Then the proteins were transferred onto a polyvinylidene difluoride membrane (Millipore, USA). After being blocked for 1 h at room temperature, the membrane was incubated with a 1:1000 dilution of rabbit polyclonal anti-mouse ET-, β-catenin, E-cadherin, RhoA and RhoC and GAPDH (ABGENT, USA) overnight. Before detected with an ECL chemiluminescence detection kit (Advansta, USA), proteins were incubated with the corresponding secondary antibody for 1 h at room temperature. The bands were obtained by GeneGnome 5 (Synoptics Ltd., UK).
Statistical analysis
All data were expressed as the mean ± SD (standard deviation) and compared using ANOVA. The T test was used for comparisons. Statistical significance was assumed as p<0.05. All statistical analyses were performed using the SPSS 13.0 statistical software.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Denmeade SR, Lin XS, Isaacs JT. Role of programmed (apoptotic) cell death during the progression and therapy for prostate cancer. Prostate 1996; 28: 251-265.
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin 2016; 66:7-30.
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069-1075.
- Jones S, Zhang X, Parsons DW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008; 321: 1801-1806.
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ. An integrated genomic analysis of human glioblastoma multiforme. Science 2008; 321: 1807-1812.
- Kim JH, Dhanasekaran SM, Mehra R, Tomlins SA, Gu W. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res 2007; 67: 8229-8239.
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 2004; 101: 811-816.
- Lieberfarb ME, Lin M, Lechpammer M. Genome-wide loss of heterozygosity analysis from laser capture microdissected pros-tate cancer using single nucleotide polymorphic allele (SNP) arrays and a novel bioinformatics platform dChipSNP. Cancer Res 2003; 63:4781-4785.
- Paik S, Shak S, Tang G, Kim C, Baker J. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004; 351: 2817-2826.
- van de Vijver, He MJ, van’t Veer YD. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347: 1999-2009.
- Febbo PG, Sellers WR. Use of expression analysis to predict outcome after radical prostatectomy. J Urol 2003; 170: S11-19.
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002; 1: 203-209.
- Patel JC, Maughan BL, Agarwal AM, Batten JA, Zhang TY. Emerging molecularly targeted therapies in castration refractory prostate cancer. Prostate Cancer 2013; 2013: 981684.
- Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature 1988; 332: 411-415.
- Xu D, Emoto N, Giaid A, Slaughter C, Kaw S. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell 1994; 78: 473-485.
- Rubin SA, Levin ER. Clinical review 53: The endocrinology of vasoactive peptides: synthesis to function. J Clin Endocrinol Metab 1994; 78: 6-10.
- Burkhardt M, Barton M, Shaw SG. Receptor- and non-receptor-mediated clearance of big-endothelin and endothelin-1: differential effects of acute and chronic ETA receptor blockade. J Hypertens 2000; 18: 273-279.
- Bremnes T, Paasche JD, Mehlum A, Sandberg C, Bremnes B. Regulation and intracellular trafficking pathways of the endothelin receptors. J Biol Chem 2000; 275: 17596-17604.
- Battistini B, D'Orléans-Juste P, Sirois P. Endothelins: circulating plasma levels and presence in other biologic fluids. Lab Invest 1993; 68: 600-628.
- Le Brun G, Aubin P, Soliman H, Ropiquet F, Villette JM. Upregulation of endothelin 1 and its precursor by IL-1beta, TNF-alpha, and TGF-beta in the PC3 human prostate cancer cell line. Cytokine 1999; 11: 157-162.
- Lees DM, Pallikaros Z, Corder R. The p55 tumor necrosis factor receptor (CD120a) induces endothelin-1 synthesis in endothelial and epithelial cells. Eur J Pharmacol 2000; 390: 89-94.
- Goldie RG. Endothelins in health and disease: an overview. Clin Exp Pharmacol Physiol 1999; 26: 145-148.
- Rubanyi GM, Polokoff MA. Endothelins: molecular biology, biochemistry, pharmacology, physiology and patho-physiology. Pharmacol Rev 1994; 46: 328.
- Mortensen LH. Endothelin and the central and peripheral nervous systems: a decade of endothelin research. Clin Exp Pharmacol Physiol 1999; 26: 980-984.
- Chiao JW, Moonga BS, Yang YM, Kancherla R, Mittelman A. Endothelin-1 from prostate cancer cells is enhanced by bone contact which blocks osteoclastic bone resorption. Br J Cancer 2000; 83: 360-365.
- Sullivan ME, Mumtaz FH, Khan MA, Dashwood MR, Thompson CS. Endothelins in the urinary tract. BJU Int 2000; 86: 97-106.
- Levy AP, Levy NS, Wegner S, Goldberg MA. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J Biol Chem 1995; 270: 13333-13340.
- Masaki T, Miwa S, Sawamura T, Ninomiya H, Okamoto Y. Subcellular mechanisms of endothelin action in vascular system. Eur J Pharmacol 1999; 375: 133-138.
- Nelson JB, Carducci M. The role of the endothelin axis in prostate cancer. Prostate J 1999; 1: 126-130.
- Nelson JB, Chan-Tack K, Hedican SP, Magnuson SR, Opgenorth TJ. Endothelin-1 production and decreased endothelin B receptor expression in advanced prostate cancer. Cancer Res 1996; 56: 663-668.
- Nelson JB, Hedican SP, George DJ, Reddi AH, Piantadosi S. Identification of endothelin-1 in the pathophysiology of metastatic adenocarcinoma of the prostate. Nat Med 1995; 1: 944-949.
- Nelson JB, Lee WH, Nguyen SH, Jarrard DF, Brooks JD. Methylation of the 5' CpG island of the endothelin B receptor gene is common in human prostate cancer. Cancer Res 1997; 57: 35-37.
- Wu-Wong JR, Chiou WJ, Dickinson R, Opgenorth TJ. Endothelin attenuates apoptosis in human smooth muscle cells. Biochem J 1997; 328: 733-737.
- Gordon C, Petit F, Kroisel P. Mutations in Endothelin 1 Cause Recessive Auriculocondylar Syndrome and Dominant Isolated Question-Mark Ears. American Journal of Human Genetics 2013; 93:1118-25.
- Kochva E, Viljoen CC, Botes DP. A new type of toxin in the venom of snakes of the genus Atractaspis (Atractaspidinae). Toxicon 1982; 20: 581-592.
- Hung VK, Yeung PK, Lai AK. Selective astrocytic endothelin-1 overexpression contributes to dementia associated with ischemic stroke by exaggerating astrocyte-derived amyloid secretion. J Cereb Blood Flow & Metab 2015; 35.
- Casey ML, Byrd W, MacDonald PC. Massive amounts of immunoreactive endothelin in human seminal fluid. J Clin Endocrinol Metab 1992; 74: 223-225.
- Langenstroer P, Tang R, Shapiro E, Divish B, Opgenorth T. Endothelin-1 in the human prostate: tissue levels, source of production and isometric tension studies. J Urol 1993; 150: 495-499.
- Walden PD, Ittmann M, Monaco ME. Endothelin-1 production and agonist activities in cultured prostate-derived cells: implications for regulation of endothelin bioactivity and bioavailability in prostatic hyperplasia. Prostate 1998; 34: 241.
- Daub H, Weiss FU, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature 1996; 379: 557-560.
- Wasilenko WJ, Cooper J, Palad AJ, Somers KD, Blackmore PF. Calcium signaling in prostate cancer cells: evidence for multiple receptors and enhanced sensitivity to bombesin/GRP. Prostate 1997; 30: 167-173.
- Kobayashi S, Tang R, Wang B, Opgenorth T, Stein E. Localization of endothelin receptors in the human prostate. J Urol 1994; 151: 763-766.