Research Article - Environmental Risk Assessment and Remediation (2017) Volume 1, Issue 1
Halotolerant Bacterial Consortium able to Degrade Substituted Phenolic Compounds Isolated from Saline Environment.
Ashwini Murthy and Krishnaswamy Veena Gayathri*
Department of Biotechnology, Stella Maris College, Chennai - 87, Tamilnadu, India
- *Corresponding Author:
- Krishnaswamy Veena Gayathri
Department of Biotechnology
Stella Maris College
Chennai - 87
Tamilnadu, India
Tel: +91 9940412363
E-mail: veenagayathri@yahoo.com
Accepted date: January 18, 2017
Abstract
Phenolic compounds are considered to be toxic compounds produced from many industrial effluents. Presence of these recalcitrant compounds in the environment posses significant health risks to humans. The substituted phenolic compounds are carcinogenic and toxic environmental pollutants which are massively discharged into the environment from anthropogenic activities. This present work focuses on the degradation of different concentrations of substituted phenolic compounds by the halotolerant bacterial consortium isolated from a saline environment. The isolated halotolerant bacterial consortium was able to grow from a primary phenolic compound like catechol to a most recalcitrant compound 2,4,6-trichlorophenol. The result showed that the moderately halotolerant bacterial consortium was able to utilize up to 250 mg/L of Catechol (55%) to 2,4,6- TCP (77%) , respectively. The isolated bacterial consortium consisted of three bacterial strains which co-existed during degradation. The16s rRNA sequencing results showed that the bacterial strains were Exiguobacterium arabatum strain YCY17 (AS1), Bacillus cereus strain I50 (AS2), and Bacillus pumilus MM15 (AS3). Such isolated bacterial consortium would be of great importance in the treatment of wastewater containing chlorophenols in the presence of salt content.
Keywords
Halotolerant bacterial consortium, Phenolic compounds, Saline environment, Wastewater, Xenobiotics,Chlorophenols.
Introduction
Environmental pollution due to anthropogenic activity has spread to all types of ecosystems. Hence, marine, freshwater, soils and air have been impacted by the dispersion of contamination. The release of chemicals into environment due to agricultural and industrial activities often creates toxicological problems as a result of they cause the pollution.
Phenol is the most common contaminant present in industrial wastewater that exhibits the toxicity which affects the living organisms such as microorganism, aquatic flora and fauna including human beings [1,2]. Phenol is an aromatic compound which is also known as carbolic acid. It is a key chemical in dyes, detergents, pharmaceutical drugs, etc. [3]. According to American Environmental Protection Agency (EPA) phenol is considered as a highly toxic pollutant [4]. The toxic level of phenol varies between the concentrations of 9-25 mg/L for fish and the concentration of 10-24 mg/L for human beings. Phenol is highly corrosive and poisoning agent that causes harmful side effects such as impaired vision, carboluria, diarrhea and mouth sores [5].
Biodegradation is defined as the metabolic ability of microorganisms to transfer organic pollutants into the less harmful and non-hazardous substances. Biodegradation process is affected by several factors such as oxygen, pH value, nutrients, concentration, composition and physical and chemical characteristics [6]. Several methods have been followed for the degradation of phenolics such as precipitation, coagulation, ion exchange, ultra filtration and the by-products from the processes [7]. The most efficient and economical way to degrade phenolic compounds present in industrial wastewater is by microorganisms [8], which degrades them in an ecofriendly way. Different microbes including bacteria, fungi and yeast have been used for the degradation phenol from different sources. Amongst the microorganisms, bacteria were considered crucial as they could degrade efficiently than the other microbes [9]. Mesophiles could degrade the phenolic compounds at low concentrations, whereas it also had many inhibitory effects significantly on wastewater treatment [10].
Different industrial activities of saline and hyper-saline environment actively contaminate the environment with organic compounds which results in environmental pollution. For the removal of these organic contaminants, biodegradation is the primary mechanism employed; however, the process is challenging to perform because of the saline conditions [6].
Halotolerant organisms are a group of microbes that could grow under saline conditions, although, they do not require high concentrations of salt for their growth. This process of surviving at salt concentrations higher than the necessary for growth is termed as halotolerance. When the growth is more than 2.5 M salt concentration it is said to be extremely halotolerant. These halotolerant organisms could be found in saline wastewater and soils that are inhabited by halophilic groups. When salinity is high, a traditional pollutant biodegradation does not function; therefore halotolerant bacterial consortium which is adapted to live in such saline conditions could be used as an alternative for the treatment of wastewater in industries for effective biological treatment of phenolic compounds [11].
Hence this study was on the degradation substituted phenolic compounds by a moderately halotolerant bacterial consortium isolated from saline environment. An isolated bacterial consortium that was enriched from saline environment, could grow on different substituted chlorophenols at different concentrations. The growth conditions such as pH, temperature, carbon and nitrogen sources of the bacterial consortium were optimized for maximum degradation of substituted phenolics. The individual bacterial strains present in the consortium were biochemically characterized and identified by 16s rRNA sequencing.
Materials and Methods
Soil sampling
Soil samples were collected from different saline sites such as salt pans, salterns in Chennai, Nagercoil and Tuticorin located in the coastal areas of Tamil Nadu. The soil samples were transferred to sterile zip lock covers and transported immediately to the laboratory. To obtain the enriched bacterial consortium, the soil samples (300 g wet weight) were added to sterile distilled water (1:1 w/v) for 1 h at room temperature and kept for shaking.
At the initial acclimatization stage, the bacterial consortium was enriched with the basic aromatic compound phenol at a concentration of 50 mg/L.
Enrichment of bacterial consortium
Bacterial consortium isolated from saline environment was initially acclimatized and enriched with phenol as the sole carbon source of energy. There were about three bacterial strains present in the consortium. Bacterial consortium was allowed to grow with different substituted phenols like Catechol, Hydroquinone and 2,4,6-trichlorophenol (2,4,6- TCP) from 25 mg/L to 100 mg/L concentrations, with the mineral salts medium of (g/L) NaCl 50.0, KH2PO4 0.25, NH4Cl 1.0, Na2BO7 2.0, FeCl3 0.0125, CaCl2 0.06 and MgCl2 0.05 with 10 mg/L of yeast extract, adjusted to pH -7 and distilled water – 1 L [12]. The medium was autoclaved, cooled down to room temperature and then was amended with Catechol, Hydroquinone and 2,4,6-TCP through a sterile filter (0.45 μm) in 250 ml Erlenmeyer flasks. The mineral salts medium contained varied NaCl concentrations ranging from 30 g/L, 50 g/L, 70 g/L and 90 g/L. The culture was then transferred, four to five times after their growth phase. Increase in viable cell count of the bacterial consortium from 105 to 106 cfu/mL with phenol was taken as a confirmation for the utilization of phenol. Once the consortium was fully acclimatized to phenol, its ability to grow on substituted chlorophenols was analysed. The chemicals and reagents used in the study were (Analar grade) obtained from Merck, India.
Identification of the Bacterial Strains in the Consortium
Biochemical characterization
Biochemical tests were examined by performing the procedure of Gram staining, Motility tests, Triple sugar iron agar, Catalase, Oxidase, Urease, Indole, Methyl red, Voges Proskauer and Citrate utilization and various sugar utilization tests. After 24 h incubation by inoculation with the respective bacterial strain at 37oC, the colour change observed and accounted for positive/ negative result. The unknown bacterial strain was studied for genus level identification which was accomplished by using Bergey’s Manual of Systematic Bacteriology [13].
16S rRNA partial gene sequencing
Chromosomal DNA was isolated from pure strains of the consortium by the standard phenol/chloroform extraction method [14]. The 1.5 kb partial sequence of 16S rDNA gene was amplified by the chromosomal DNA using poly-merase chain reaction (PCR) with universal Eubacteria-specific primers 16F27 (5-CCA GAG TTT GAT CMT GGC TCA G-3) and 16R1525XP (5-TTCTGCAGT CTA GAA GGA GGT GWT CCAGCC-3) [15]. The PCR amplification conditions were: initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 10 min; and finally sequencing was performed on an ABI310- automatedDNA sequencer using Big Dye terminator kit (Applied Biosystems 3730x l DNA Analyzer). The amplified 16S rRNA gene (PCR products) from these isolates was directly sequenced after purification by precipitation with polyethylene glycol and NaCl. The primers used to obtain the complete sequence of 16S rRNA gene of the isolates were the same as used for the PCR amplification (16F27N and 16R1525XP). Sequence analysis was performed using Chromas Prosequence analysis software. The phylogenetic tree was constructed by MEGA6 software.
Degradation of substituted phenols
For the batch study on degradation, mineral salts mediums containing Catechol, hydroquinone and 2,4,6-TCP were inoculated with the bacterial consortium. The different conditions used for the degradation of substituted phenols were (i) mineral salts medium+substituted phenols+bacterial consortium; (ii) mineral salts medium+substituted phenols; and (iii) mineral salts medium+bacterial consortium; with (ii) and (iii) serving as controls. The bacterial consortium was added along with the medium at concentrations of 104-105 cfu/mL. The culture was incubated at 37ºC with shaking at 150 g and extracted for every 24 h interval for 5 days. Increase in protein yield was considered as the ability of the bacterial consortium to degrade the substituted phenols. Substituted phenolic compounds were added to the medium at different concentrations (50 mg/L to 250 mg/L). Bacterial Cell morphology and the motility of cells were examined by light microscopy. The viable cell count (cfu/ mL) was also counted by plating the cells on nutrient agar.
Effect of pH, salt, carbon and nitrogen sources
Effect of different pH (5.5, 6, 6.5, 7, 7.5, 8 and 8.5) on the substituted phenol degradation was studied with the bacterial consortium at a optimum concentration of 100 mg/L 2,4,6-TCP at 5% NaCl. Effects of different concentrations of NaCl were studied from 3%, 7% and 9% in the mineral salts medium. The mineral salts medium was substituted with different carbon sources at a concentration of 0.5% using glucose, fructose, lactose, and sucrose. Effect of different nitrogen sources on the removal of chloro phenolic compounds was determined by adding of 0.5% of KNO3, NH4NO3, NH4SO4 and NaNO3 to the mineral salts medium.
Gas chromatographic analysis of phenolic compound degradation
The ability of the consortium to utilize phenol as sole carbon source was determined by growing it in the mineral medium containing different concentrations of phenol from 50 mg/L to 250 mg/L. The cell suspensions were clarified by centrifugation at (10,000 rpm for 15 min, 6°C). The culture supernatant was extracted with dichloromethane and filtered through a 0.2 μm Gelman filter acro disc, prior to analysis in gas chromatograph (Chemito GC Model No 1000) equipped with FID detector and capillary column(Varian Chromopak capillary column CP SIL 8 CB, 30 m × 0.32 mm with detection limit of 10 ppt of the compound. Nitrogen was used as a carrier gas, injector temperature was 220°C, detector temperature was 250°C and the oven temperature of the column was maintained at 150°C. Standard solutions of different phenolic compounds were used for reference. The samples were injected one by one and the utilization rates of phenolic compounds were calculated based on the peak area percent and retention time.
Results and Discussion
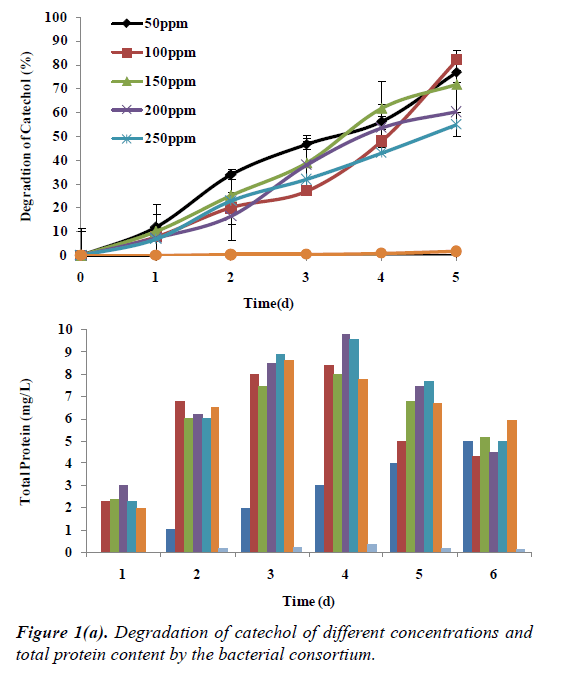
Recalcitrant phenolic compounds recalcitrant compounds which are main constituents of many industrial effluents. For the effective removal of these compounds a suitable system has to develop for safely treating and disposing the phenolic compound containing effluents. In the present study soil samples were collected from the salterns were enriched with phenolic compounds to isolate halotolerant bacterial consortium that could utilize all the phenolic compounds used in the study. During the isolation period, several bacterial strains which could degrade substituted phenolic compounds under saline conditions survived in the consortium. After successive transfer during the enrichment period, only three bacterial strains coexisted and were named as (AS1, AS2, AS3). These could survive and utilize substituted phenolic compounds as a sole source of carbon and energy. Growth of the bacterial consortium during the enrichment with catechol as sole carbon source was studied it showed that there was an increase in viable cell count (10-5 × 108 cfu/ml) till 24 h and decrease in viable cell count (10-8 cfu/ml) indicating that bacterial consortium could survive in the presence of catechol up to 96 h. Hence for all the further experiments on the growth and degradation of the substituted phenolic compounds were studied up to 96 h.
García et al. studied the degradation of low- molecular- weight aromatic compounds (benzoic acid, p-hydroxy benzoic acid, cinnamic acid, phenylacetic acid, p-coumaric acid, ferulic acid, salicylic acid) by a group of halophilic bacteria. When the isolates were enriched on phenol, they could utilize a greater number of aromatic compounds than the rest of the isolates enriched on other aromatic compounds other than phenol, which showed wider substrate specificity. But the mixed halophilic isolates were unable to utilize p-Cresol. In the present work the isolated halotolerant bacterial consortium was able to grow with all the substituted chlorophenols under saline conditions which are higher molecular weight compounds than the low molecular weight compounds used in the previous literature study.
Degradation of substituted phenols at different concentrations
The ability of the bacterial consortium to utilize catechol as sole source of carbon and energy was studied at different concentrations of 50 mg/L to 250 mg/L (50 mg/L, 100 mg/L, 150 mg/L, 200 mg/L and 250 mg/L), respectively. Catechol at 250 mg/L concentration showed degradation up to 55% by the end of 5th day. This was followed by 150 mg/L of catechol concentration showing degradation up to 60% by the end of 5th day. When the concentration was increased to 200 mg/L, there was a reduction in the removal percentage of catechol to 60%, at 100 mg/L it showed a maximum removal of 82% and at 50 mg/L a degradation of 77%, respectively. There was a corresponding decrease in the growth of bacterial consortium represented by the protein content as predicted in the Figure 1a.
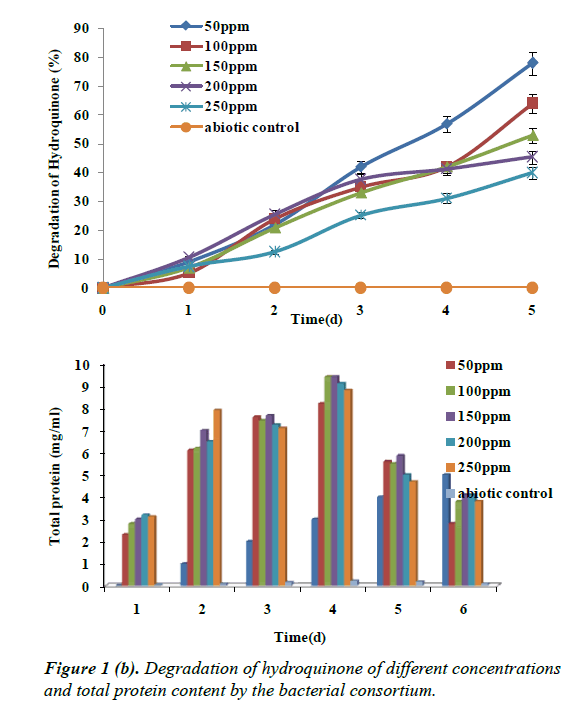
Hydroquinone at 250 mg/L concentration showed degradation up to 40% by the end of 96 h, this was followed by 200 mg/L of hydroquinone concentration showing degradation of 45.5%, 150 mg/L showing degradation up to 53%, 100 mg/L showed degradation up to 64%, and 50 mg/L showing a maximum degradation up to 78% which is showing in the Figure 1b.
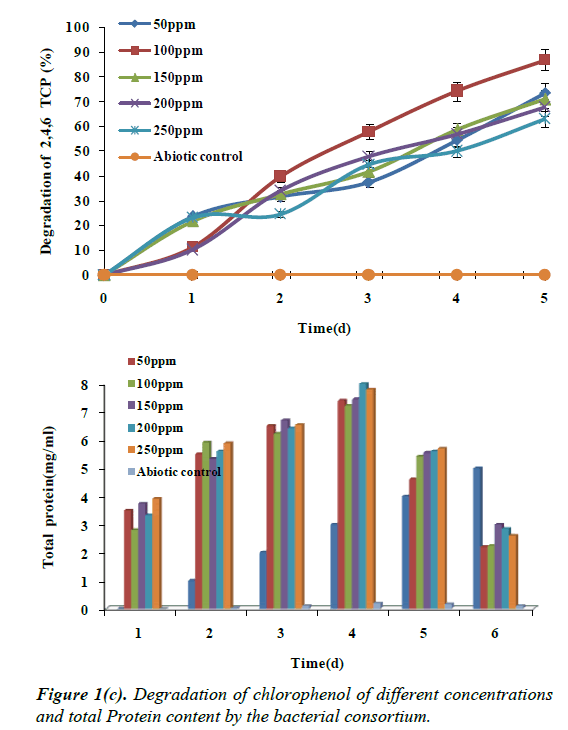
2,4,6-TCP at 250 mg/L of concentration showed degradation up to 63% by the end of 96 h and a maximum growth during 48 h of incubation. This was followed by 200 mg/L of 2,4,6-TCP concentration showing increase in degradation up to 67% by the end of 96 h for 150 mg/L, there was reduction in the degradation to 71%, and there was corresponding decrease in the growth of the bacterial consortium. The degradation of 2,4,6-TCP was maximum at 100 mg/L concentration showing 87% , followed by 50 mg/L of about 73% , which is depicted in the Figure 1c. To our knowledge there are not much reports on the degradation of 2,4,6-TCP under saline conditions. However, biodegradation of chlorophenols by a variety of microorganisms have been studied by many authors [17] under non-saline conditions. According to these studies, the biodegradability of chlorophenols depends on the number and position of halogens in the aromatic ring. Furthermore, high chlorophenol concentrations are known to be inhibitory to microbial growth.
Kharoune et al. reported the degradation of 2,4,6-TCP (200 mg/L) under non-saline conditions by a microbial consortium containing Sphingomonas paucimobilis, Burkholderia cepacia, Chryseomonas luteola and Vibrio metschnikovii, where only about 51 mg of 2,4,6-TCP g/L cell protein/h was degraded in 160 h. Pamukoglu et al. reported that the degradation of 2,4,6- TCP under non-saline conditions by Rhodococcus rhodochrous (DSM 43241) where 150 mg/L of 2,4,6-TCP inhibited the growth and degradation, in the presence of glucose as the additional carbon source. The bacterial consortium effectively degraded mono- and di-chlorophenols when these were used as the only sources of carbon and energy. The biodegradability of the compounds decreased in the order: 4-CP >2,4-DCP >2,4,6- TCP. In chlorophenol biodegradation studies, it can be noted that 4-CP, 2,4-DCP were degraded readily than 2,4,6-TCP.
Optimization of Growth Conditions on the Degradation by the Bacterial Consortium
Effect of pH on the degradation of 2,4,6-TCP
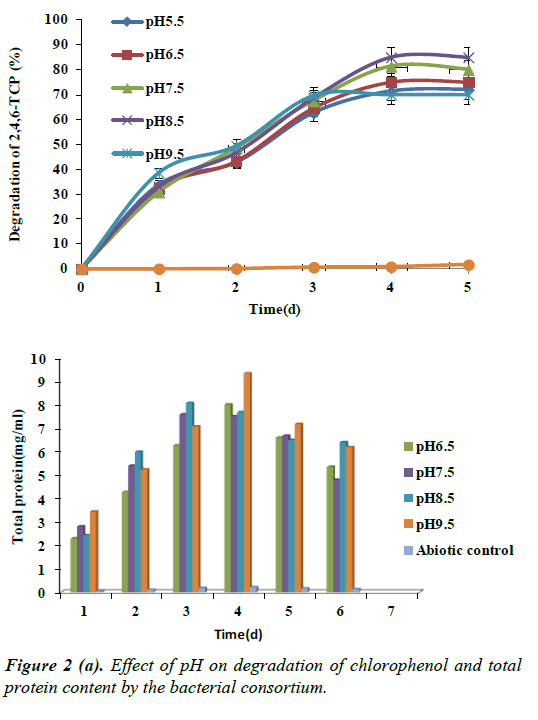
Effect of pH on the degradation of 2,4,6-TCP was studied at 5% NaCl at optimum concentration of 100 mg/L of chlorophenol. Figure 2 shows the growth and degradation of 2,4,6-TCP by bacterial consortium at different pH levels. At pH 5.5 the degradation of chlorophenol was up to 72%, which increased with pH 6.5 to 75% and a maximum of 85% at pH 8.5. The degradation of 2,4,6-TCP reduced to 70% with pH 9.5 by the end of 96 h. According to research work by García et al. the pH values above 6.0 were suitable for all the strains; However growth at pH 7.5 was found to be optimal. None of the isolates exhibited growth at pH 4.5. The pH of the medium plays a vital role in most of the microbial processes. In the present study halotolarent bacterial consortium which was isolated from saline environment could degrade substituted phenol compounds up to a pH 9 at optimum temperature of 36ºC. This shows that the halophilic nature of bacterial consortium was able to grow even at alkaline pH.
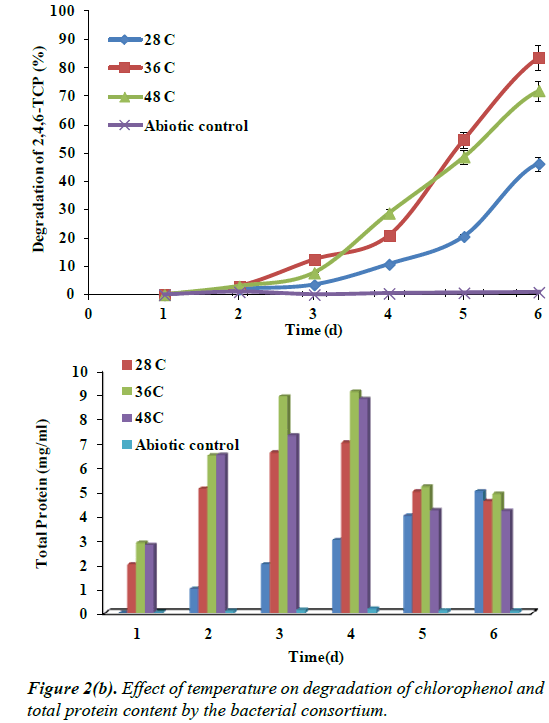
Effect of Temperature on the degradation of 2,4,6- TCP
The study was carried out to determine the effect of temperature (28ºC, 36ºC, 48ºC) on the degradation of at optimum concentration of 2,4,6-TCP 100 mg/L. Figure 2b shows the growth and degradation of 2,4,6-TCP at different temperature. The degradation of 2,4,6 TCP being maximum at 36ºC showing degradation up to 89% by the end of 96 h. At 28ºC and 48ºC the degradation of chlorophenol showed only up to 40% and 72%, removal respectively.
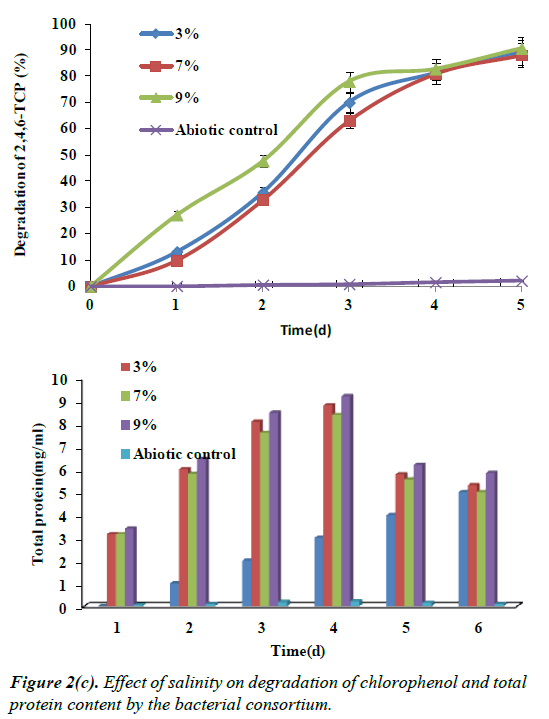
Effect of salinity on the degradation of 2,4,6- TCP
Salinity plays an important role in the metabolism of halophilic bacteria; Hence the isolated halotolerant bacterial consortium was also checked with different concentration of NaCl. Growth and degradation of 2,4,6-TCP (100 mg/L) was studied in three saline concentrations i.e. 3%, 7% and 9%, in which 3% of NaCl concentrations showed maximum removal of 90%, and there was also simultaneous increase in the growth showing maximum growth, which predicts the halotolerance of the isolated bacterial consortium. At 7% and 9% NaCl degradation reached up to 85% and 72% as shown in the Figure 2c. In this study, growth and degradation potential of the isolated bacterial consortium was checked with different NaCl concentrations. It could be depicted from the figure that 3% NaCl concentration showed maximum removal than the other concentrations used.
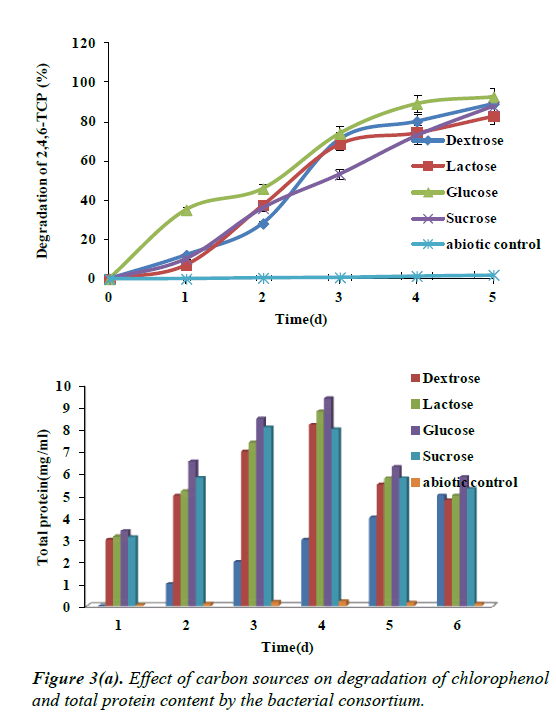
Effect of carbon sources on the degradation of 2,4,6- TCP
To examine the influence of carbon sources on the degradation of 2,4,6-TCP at optimum concentration of 100 mg/L, carbon sources like glucose, dextrose, sucrose, lactose were supplemented along with 2,4,6-TCP. Almost all the carbon sources were able to enhance the degradation of 2,4,6-TCP. A great advantage for this degradation ability was particularly observed in the presence of glucose as carbon source showing degradation up to 92%. This was followed by sucrose, dextrose and lactose having 87%, 89% and 82% of degradation as shown in the Figure 3a. Hence maximum growth was observed with glucose as carbon source supplemented in the medium.
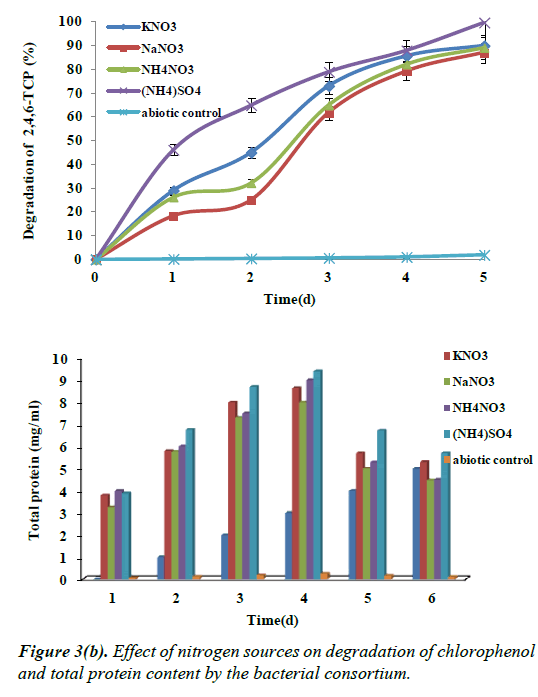
Effect of nitrogen sources on the degradation of 2,4,6- TCP
To examine the influence of nitrogen sources on the degradation of 2,4,6-TCP at optimum concentration of 100 mg/L, various nitrogen sources like ammonium sulphate, ammonium nitrate. Potassium nitrate and sodium nitrate were supplemented along with chlorophenol. Figure 3b shows that ammonium sulphate, ammonium nitrate, potassium nitrate and sodium nitrate played an important role in enhancing the growth and degradation of chlorophenol, ammonium sulphate being the maximum showing degradation up to 99% respectively by the end of 96 h. This was followed by ammonium nitrate, sodium nitrate and potassium nitrate showing 89%, 87% and 90% of degradation respectively by the end of 96 h.
Biochemical and molecular characterization of the bacterial isolates
The bacterial consortium consisted of three bacterial strains, which were all gram positive. Table 1 summarizes the results on the biochemical tests of the isolated bacterial strains. The biochemical characteristics of three bacterial strains showed that they belong to the phyla Firmicutes. These groups of bacteria are commonly present in the hypersaline environments. All the three strains were round in shape with smooth and had irregular morphology on the nutrient agar plate. AS1 showed translucent raised colony with irregular edges. AS2 was small transparent flat colony with smooth edges and AS3 was big yellow mucoid colony with smooth edges. Table 2 gives the Antibiotic Susceptibility test for the three bacterial isolates. Through 16s rRNA sequencing, the bacterial strains were identified as Exiguobacterium arabatum strain YCY17 (AS1)-accession number KX608938, Bacillus cereus strain I50 (AS2)-accession number KX608939, and Bacillus pumilus MM15 (AS3)-accession number KX608940. Table 3 shows the phylogenetic affiliation of the bacterial isolates. Figure 4 predicts the Phylogenetic affiliation between the three bacterial strains present in the consortium [20].
| CHARACTERISTICS | AS1 | AS2 | AS3 |
|---|---|---|---|
| Gram staining | + | + | + |
| Motility | + | + | + |
| Triple sugar iron agar | + | + | + |
| Catalase | + | - | + |
| Urease | - | - | - |
| Indole | + | - | - |
| Methyl red | - | - | + |
| Voges proskauer | - | - | - |
| Citrate utilization | + | + | + |
Table 1. Biochemical characteristics of the isolated bacterial strains.
| S.NO | Antibiotic Disc | ZONE OF INHIBITION DIAMETER (cm) | ||
|---|---|---|---|---|
| AM1 | AM2 | AM3 | ||
| 1 | Ampicillin(10 µg/disc) | 3 (S) | - (R) | -(R) |
| 2 | Azithromycin(15 µg/disc) | 3(S) | 3 (S) | 3 (S) |
| 3 | Amoxycillin(10 µg/disc) | 3 (S) | - (R) | - (R) |
| 4 | Co-Trimaxazole(25 µg/disc) | 3 (S) | 3 (S) | - (R) |
| 5 | Erythromysin(15 µg/disc) | 3 (S) | 3 (S) | 3 (S) |
| 6 | Gentamycin(10 µg/disc) | 3 (S) | 2.5 (S) | 3(S) |
| 7 | Penicillin G(10 µg/disc) | - (R) | -(R) | - (R) |
| 8 | Tetracycline(30 µg/disc) | 3 (S) | 2.5(S) | - (R) |
NOTE: S=Sensitive; R=Resistance; -=No zone of inhibition; I=Intermediate.
Table 2. Antibiotic Susceptibility test for the three bacterial isolates.
| Isolate No. | Phylum | Nearest phylogenetic neighbour (Accession no.) | Affiliation (Accession no.) |
Similarity (%) |
|---|---|---|---|---|
| AS1 | Firmicutes | Exiguobacterium arabatumstrain YCY17 (KF560366.1) | Exiguobacterium arabatum KX608938 |
99 |
| AS2 | Firmicutes | Bacillus cereusstrain I50 (KC700352.1) | Bacillus cereus KX608939 |
99 |
| AS3 | Firmicutes | Bacillus pumilus MM15 (JX083963.1) | Bacillus pumilus KX608940 |
99 |
Table 3. Bacterial strains identified by 16s r-RNA sequencing.
Conclusion
In the present study, it could be noted that the isolated bacterial consortium were able to utilize all the substituted phenolic compounds like catechol, hydroquinone and 2,4,6-Trichlorophenol which proves the consortium had the ability to degrade different range of phenolic compounds from mono to Trichlorophenol under saline conditions. The obtained results showed that the increase in the concentration of the substrate reduced the growth of the consortium which was also represented by the protein yield. When the concentration of the substrate increased to 100 mg/L the degradation reduced to 74%, which might be due to toxicity of the substrate or unavailability of 2,4,6-TCP to the consortium or accumulation of toxic metabolites or depletion of nutrients such as nitrogen and phosphate or all these factors together would have inhibited the growth of the bacterial consortium. Another possible reason would be other than that the substrate no other carbon source like glucose was supplied in the medium [21-23].
The results from the present study have demonstrated the ability of halotolarent bacterial consortium to rapidly degrade substituted phenolic compounds at different salt concentrations where the optimum degradation was achieved at 5% NaCl under aerobic conditions. Therefore, isolated bacterial consortium could be applied in the treatment of soil or wastewater contaminated with phenolic compounds. Thus, these experimental results provide hope for the establishment of cost effective methods to remediate phenolic wastewater effluents with high salt content.
Acknowledgment
The authors would like to thank the Management of Stella Maris College for providing them the opportunity to perform research and financial assistance.
References
- Micha?owicz J, Duda W. Phenols- sources and toxicity.Polish J Environmen Studies. 2007;16:347-62.
- Veena Gayathri K, Namasivayam V. Enrichment of phenol degrading moderately halophilic bacterial consortium from saline environment.J Biorem Biodeg. 2010;1:104.
- Nawawi, Norazah Mohammad, et al. Statistical optimisation for improvement of phenol degradation by Rhodococcus sp. NAM 81.J of Env Biol.2016;37:443.
- Rao NN. Liquid-liquid extraction of phenol from simulated sebacic acid wastewater.J Sci Ind Res.2009;68:823-28.
- Kulkarni SJ, KawareJP. Review on research for removal of phenol from wastewater.Int J Sci and Res Publi. 2013;3:1-4.
- Margesin R, Schinner F. Biodegradation and bioremediation of hydrocarbons in extreme environments.Applied Microbiol Biotechnol. 2001;56:650-63.
- Rengaraj S, Moon SH, Sivabalan R. Agricultural solid waste for the removal of organics: adsorption of phenol from water and wastewater by palm seed coat activated carbon. Waste Management. 2002;22:543-48.
- Sivasubramanian S, Karthick SRN. Optimization of parameters for phenol degradation using immobilized Candida tropicalis SSK01 in batch reactor.Journal of Environmental Biology.2014;35:531.
- Kafilzadeh F, Farhangdoost MS, Tahery Y. Isolation and identification of phenol degrading bacteria from Lake Parishan and their growth kinetic assay. Afri J Biotechnol. 2010;9:6721-26.
- Singlande E. Improvement of the treatment of salted liquid waste by integrated electrodialysis upstream biological treatment.Desalination. 2006;199:64-6.
- Oren Aharon. Microbial life at high salt concentrations: phylogenetic and metabolic diversity.Saline systems. 2008;4:1.
- Alva V, Peyton BM. Phenol and catechol biodegradation by haloalkaliphile Halomonas campisalis: Influence on pH and salinity. Environment Sci Technol. 2003;37:4307-4402.
- Bergey’s Manual of Systematic Bacteriology, 2nd e. Brenner DJ, Kreig NR and Staley JT, Springer, editors. New York; 2005.
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor: NY; 1989.
- Pidiyar V, Kaznowski A, Narayan NB. Aeromonas culicicola sp nov, from the midgut of Culex quinquefasciatus. Int J Syst Evol Microbiol. 2002;52:1723-28.
- García MT. Halomonas organivorans sp nov, a moderate halophile able to degrade aromatic compounds. Int J Sys Evo Microb. 2004;54:1723-28.
- Kargi F, Eker S. Removal of 2,4-Dichlorophenol and toxicity from synthetic wastewater in a rotating perforated tube film reactor. Proc Biochem. 2005;40:205-11.
- Kharoune L, Kharoune M, Lebeault J. Aerobic degradation of 2,4,6-trichlorophenol by a microbial consortium-selection and characterization of microbial consortium. Appli Microbio Biotechnol.2002;59:112-17.
- Pamukoglu M, Yunus FK. Biodegradation kinetics of 2,4,6-trichlorophenol by Rhodococcus rhodochrous in batch culture.Enz Microbial Technol. 2008;43:43-7.
- Hauben L, Vauterin L, Swings J. Comparison of 16S ribosomal DNA sequences of all Xanthomonas species. Int J Syst Bacteriol. 1997;47:328-35.
- Veena Gayathri K, Vasudevan N. Ortho and meta cleavage dioxygenases detected during the degradation of phenolic compounds by a moderately halophilic bacterial consortium. Int Res J Microbiol. 2011;2:406-14.
- Veena GK, Vasudevan N. Degradation of Chlorophenolic compounds by a moderately halotolerant bacterial consortium isolated from saline environment. International Journal of Bioassays. 2015;4:4102-08.
- Veena GK, Vasudevan N. Degradation of dual phenolics by a moderately halophilic bacterial consortium and its degradation products. Int J of Current Microb and Appl Sci. 2015;4:1083-95.