Research Article - Biomedical Research (2017) Volume 28, Issue 7
Fusion of local and global features: A proficient advent for brain tumor classification with PSO and ELM
Josephine Sutha V1* and Latha P2
1Sardar Raja College of Engineering, Tirunelveli, India
2Government College of Engineering, Tirunelveli, India
Accepted date: December 9, 2016
Abstract
Medical diagnostics is an important task to think about the inner structures and functions of human body and can be done through the investigation of digital imaging system. At the outset, detection of brain tumors is somewhat difficult in the diagnostic procedure. Magnetic Resonance Imaging (MRI) technique is a vibrant imaging method to locate the tissue contrast for anatomical details and to investigate the brain tumor locations. Contrast medium is the one frequently given before going for MRI or CT scans and is a matter that is absorbed or added into the veins to illuminate tumors. Consequently, at occasional times the image produced by the MR imaging test tends to go for illumination changes which shrinks the accuracy of automatic brain tumor detection scheme. Henceforth, in this work, GSIFT (Geometric Scale Invariant Terrain Feature Transform) is introduced as it has major progressions under blur and illumination fluctuations. Feature extraction of the proposed work is carried out by computing local features (GSIFT) and global features. Besides modified Particle Swarm Optimization (PSO) selection algorithm is applied for dimensionality reduction with the Extreme Learning Machine (ELM) Classification algorithm. The comparison ELM classifier assessment with and without PSO algorithms proves that the classification accuracy is higher in classification with PSO than the other. The proposed work is intended for automatic brain tumor detection and classification.
Keywords
Magnetic resonance imaging, Extreme learning machine, Particle swarm optimization, GSIFT (Geometric Scale Invariant Terrain Feature Transform).
Introduction
Brain is the smallest and paramount enchanting organ in human body. Scientists and academics have conferred that the human brain is the basis for sensation, remembrance, reasoning and awareness of the physical body. All parts of the body except brain can live for a while after death. But doctors confirm the death of human body once the brain got dead. Therefore everyone gives utmost preference to brain and the brain diseases. As a consequence, once the brain is disturbed by brain tumor it affects all the related activities of the human body. Early detection of brain disease and immediate medication can prevent the brain from the widespread annihilation. So it is inevitable to find out the brain tumor at its early stage. Patients must to seek right treatment immediately in order to avoid further brain disturbances.
Finding out the brain tumor at its early stage is a tedious task and a time consuming process for the rural people. There are many procrastinating factors affect the rural people not seeking immediate medical advice. These factors may be the reasons for the increase of mortality rates in rural areas. The foremost adjournment for the early detection of brain tumor includes more number of tests, Choose of right Doctor, Rendezvous of doctors, Financial Implications, Second Opinion and First Opinion of the specialist [1]. Hence, the rural people requisite a computer aided system consequently their family doctor or a normal doctor can tell the status of the tumor. In India, 80% of the residents live in rural regions, while 80% of the doctors are in the big towns [2].
In literature many authors deliberated the importance of automatic brain tumor detection in the rural areas. Since no multi-hospital in the rural, simple methods to find out the brain tumor is inevitable. The proposed diagnostic method interestingly can be handled by the novice neuro-oncologist and by the family doctors as well. This can be taken as a great chance for remote and rural patients who live in outside the regional center and frequently find it difficult to make an appointment with a neuro-oncologist. The lifetime of the individual affected by the brain tumor will increase when it is recognized at earlier times [3].
The significance of diagnosing the brain tumor in the initial stage is to decrease mortality rate and can be accomplished by accurate computer-aided diagnostic methods. There are Different algorithms identified for brain tumor location still brain tumor is one of the main sources of death among societies. It is proved by different scientists that the chance of survival can be prolonged if the tumor is predicted at its initial stage. Henceforth, there is a necessity for a dynamic approach with rapid calculation time and efficient accuracy to spot earlystage tumors is a vital objective for doctors.
Materials and Methods
The proposed technique dealt with local and global feature extraction with modified Particle Swarm Optimization (PSO) selection algorithm. Besides, Extreme Learning Machine (ELM) classifier is used for the classification of brain images. The subsequent sections delineate the theoretical information about proposed classification approach, local feature extraction and global feature extraction, Fusion of Local Feature Extraction and Global Feature Extraction and the modified PSO selection algorithm.
Proposed classification approach
In this study an Extreme Learning Machine (ELM) is applied in order to classify brain tumor cells [4]. The proposed methodology is alienated into two key phases; former is training phase and the latter is testing phase. In the training phase the classifier is trained by means of the recognized image models. In the testing phase, the features of the testing image models are taken and related with the features of the trained phase models, and hence the classified output is determined with respect to the trained image models. Contrast medium is the one frequently given before going for MRI or CT scans and is a matter that is absorbed to illuminate tumors. Since, it is tough to see many tumors without contrast it becomes an essential one before going for the particular imaging test [5].
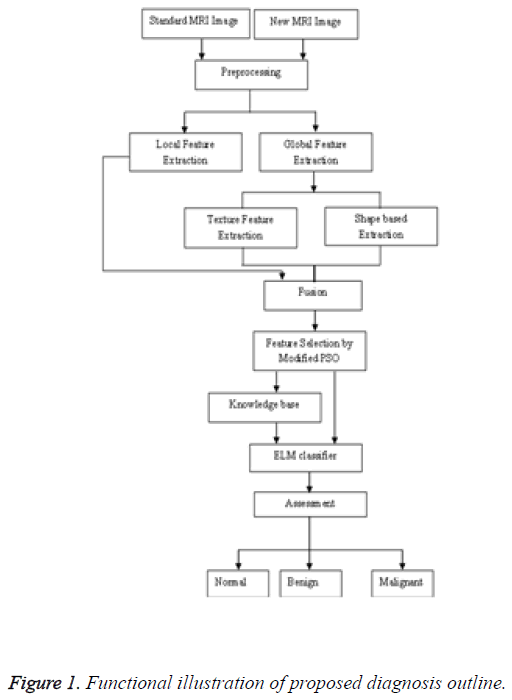
As a result, at rare times the image produced by the MR imaging test tends to go for illumination changes which reduces the accuracy of automatic brain tumor detection system. Hence, in this study for local feature extraction, SIFT descriptor with global context (GSIFT) is used, as it has significant progressions under blur and illumination fluctuations [6]. Besides Global feature extraction routine dole out with Texture based Feature Extraction, Shape based feature extraction that can differentiate the brain tumor cells based on their features [7]. Apart from that a modified particle swam optimization is selected as a feature selection algorithm [8]. The extracted features of the feature selection process were given to the ELM classifier. The ELM classifier results are assessed in to three categories normal, benign, malignant. Figure 1 expresses the functional illustration of the proposed diagnosis procedure.
Local feature extraction
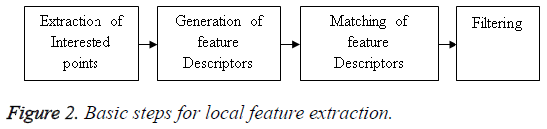
A local feature describes a portion of an image within an image. As the Local features are robust to partial occlusion, clutter, and geometric transformations they are well matched for image recognition and matching applications. Normally Scale-Invariant Feature Transform (SIFT), Principal Components Analysis (PCA)-SIFT, and Speeded up Robust Feature (SURF) are considered as a local feature detectors [9]. Among the above said descriptors SIFT and its variants have a high position among researchers. This work concentrates a variant of SIFT known as GSIFT. The necessity of GSIFT in the suggested algorithm is to incorporate a global texture vector with the descriptors generated by SIFT in the database images, as GSIFT descriptor is best for blur and illumination changes of MRI images [5]. The Basic Steps of local feature extraction is given in Figure 2.
The main hint of GSIFT is to boost global texture information, so as to give growth of curve shaped information. This GSIFT creates a vector with two parts for each spotted key point. One is the SIFT descriptor of a local feature and another part is a global texture vector practiced to distinguish related local features. Similar to SIFT, global texture (GSIFT) also produces a histogram. Moreover GSIFT needs to calculate the utmost curvature of each pixel, which is defined as the biggest utter eigenvalue of a Hessian matrix. For every key point, GSIFT creates log-polar coordinates around the key point and splits a circular area into a number of regions. Afterwards it calculates discrete values of angles and radial distances of every key point.
Global feature extraction
Global features illustrate about the whole image, in spite of the content of secluded pixels. Normally these features have an advantage that it is not suitable for spatial localization of objects as it is inflated by clutter and partial occlusion. Global feature extraction [7] has been classified as Texture based feature extraction, shape based feature extraction and histogram based feature extraction. Texture features can be extracted by structural based feature extraction, statistical based feature extraction, and model based feature extraction and transform based feature extraction. This work focuses more on statistical based feature extraction. Table 1 shows the equation of texture features taken for the diagnosis of brain tumor. In Shape based feature extraction the image is condensed to a group of shapes which can be characterized as a set of parameters and the same can be taken as set of features for processing the input image. Proposed method clarifies two shape properties which contain Area and perimeter.
| S. No | Feature | Equation |
|---|---|---|
| 1 | Energy | 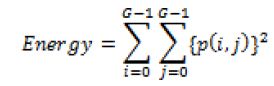 |
| 2 | Correlation | 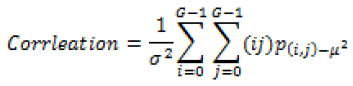 |
| 3 | Inertia | 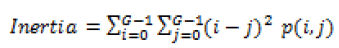 |
| 4 | Entropy | 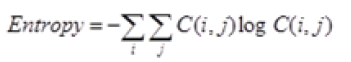 |
| 5 | Inverse difference moment |  |
| 6 | Sum average | 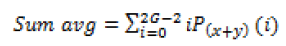 |
| 7 | Sum variance | 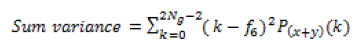 |
| 8 | Sum entropy | 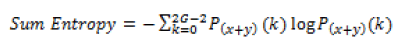 |
| 9 | Difference variance | 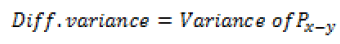 |
| 10 | Difference average | 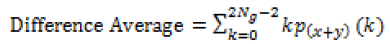 |
Table 1: Texture features taken for the diagnosis of brain tumor.
Fusion of local feature extraction and global feature extraction
Integration of different feature set is termed as feature fusion. Normally a fused feature set consists of concentrated data about the given image. Hence, fusion process yields good results. By the way when considering practically, it is very tough to accomplish feature fusion because of the following reasons:
• Several modalities may be mismatched.
• Interconnection of two or three feature sets results in large dimension taking more time to complete the fusion.
As all the above said difficulties overtake the fact of getting good efficiency, Fusion is employed in the proposed method. A technique using GSIFT with texture and shape based feature extraction is employed in this work.
Feature selection by modified particle swarm optimization (PSO)
Particle swarm optimization (PSO) is a population grounded stochastic minimization method proven by Eberhart et al. [8]. Algorithm for modified PSO is given in Figure 3. PSO prepares a group of random particles and inspect for finest particle by renovating the generations. All the particles are renovated by two “best” values in each iteration. The first one is the best fitness value and is designated as “P Best”. Another one is found by the particle swarm optimizer and is designated as global best “gBest”. Moreover when a particle has a role in the population, the best value is designated as local best “L Best”. After the identification of the two best values, the particle renovates its velocity and locations with the Equations 1 and 2.
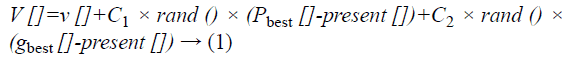
Here v [] is the particle velocity, present [] is the present particle, rand () is a random number between (0, 1). Also C1, C2 are learning factors.
Extreme Learning Machine (ELM) classification
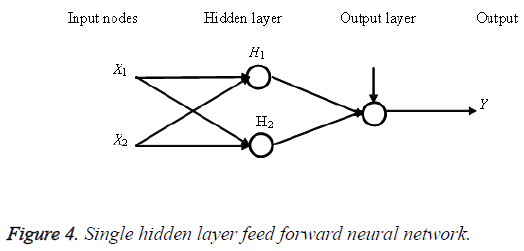
Extreme Learning Machine (ELM) is a Single Hidden Layer Feed Forward Neural Network (SLFNN) which arbitrarily chooses input weights and hidden neuron biases without training [4]. The output weight is analytically determined using least-square solution and Moore-Penrose inverse of a general linear system [10]. Thus, it permits a considerable reduced training time. Dissimilar activation functions and linear activation functions have been chosen for hidden neuron layer and for the resultant neurons. The SLFNN evaluated in the study use an additive neuron design instead of kernel based selection; hence this can be reflected as random parameter selection. The structure of ELM network is shown in Figure 4. The summarization of the ELM algorithm can be given as a three-Step Learning Model.
Let g be the activation function and M is the number of hidden nodes then for N number of inputs={(xi, ti)|xi Rn, ti Rm, i=1,… , N} the algorithm as follows:
1. Allot arbitrarily input weight vectors ai and hidden node bias bi, i=1, …, M.
2. Compute the hidden layer output matrix H.
3. Compute the output weight=H † T.
Where H† is the Moore-Penrose generalized inverse of hidden layer output matrix H [10]. By the completion of the proposed ELM algorithm, classification is meritoriously accomplished for the given testing data.
Experimental Results
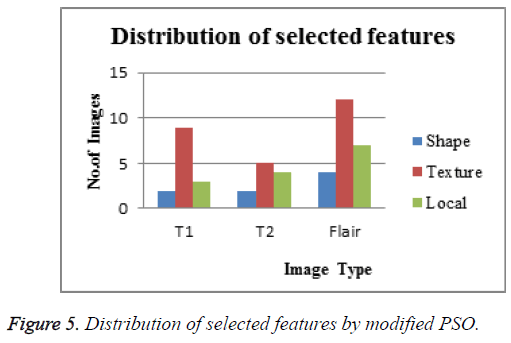
MRI scans images with the size of 512 × 512 in DICOM layout is considered in order to evaluate the performance of proposed system. The researchers used MATLAB as an environment to implement the proposed system. Proposed work is implemented with 120 normal MRI brain images and 285 abnormal MRI brain images. At first all the MRI brain images are pre-processed through wiener filter. Next GSIFT is applied for the extraction of local features (8 features) with texture based Features (10 features) and Shape based features (2 features). The obtained texture feature values are presented in Tables 2 and 3. Likewise Table 4 gives the shape based feature values. These resultant features are given together as the input for modified PSO Feature Selection algorithm. Table 5 gives the Specification of proposed PSO routine to assess the performance of modified PSO algorithm. Figure 5 shows the Distribution of selected features by modified PSO algorithm. By the way the dataset is randomly distributed as two sets with 70% as the training set and 30% as the testing set. In the training phase the outputs of modified PSO is stored as knowledge base. In the testing phase the selected features are given as input to ELM classifier. The design of extreme learning machine depends upon the number of hidden nodes of its hidden layer. Experiments were conducted by varying different hidden nodes starting from 10 and ending with 60. Outcomes of the proposed method yields maximum classification accuracy of 99% with 43 hidden nodes. The initial weights and bias of the ELM classifier is fixed between (+1, -1). Also unipolar sigmoidal activation function is found to be best activation function for the proposed system. There are two cases analysed in Table 6. One is ELM classification with modified PSO and the other is ELM classification without modified PSO. Among the two cases ELM with modified PSO produces higher accuracy than the other.
| Feature | Image l | Image 2 | Image 3 |
|---|---|---|---|
| Energy (ASAD) | 0.0000 × 102 | 0.0000 × 102 | 0.0000.1 × 102 |
| Entropy | 0.0110 × 102 | 0.0104 × 102 | 0.0115.1× 102 |
| Correlation | 0.0009 × 102 | 0.0008 × 102 | 0.0008× 102 |
| Inertia | 0.1890× 102 | 0.1657× 102 | 0.26344 × 102 |
| IDM | 0.002 × 102 | 0.0003 × 102 | 0.002 × 102 |
| Sum average | 0.1950 × 102 | 0.2010 × 102 | 0.1750 × 102 |
| Sum variance | 5.2218 × 102 | 42012× 102 | 3.8209× 102 |
| Sum entropy | 0.0087× 102 | 0.0077 × 102 | 0.0081 × 102 |
| Difference average | 0.0075 × 102 | 00091× 102 | 0.0083× 102 |
| Difference variance | 0.1167 × 102 | 0.1234× 102 | 0.2753 × 102 |
Table 2: Texture features of three malignant tumors from the database.
| Feature | Image l | Image 2 | Image 3 |
|---|---|---|---|
| Energy (ASAD) | 0.0000 × 104 | 0.0001 × 104 | 0.0000 × 104 |
| Entropy | 0.011 × 104 | 0.0011 × 104 | 0.0119 × 104 |
| Correlation | 0.0001 × 104 | 0.0002 × 104 | 0.0002 × 104 |
| Inertia | 0.0312 × 104 | 0.0412 × 104 | 0.0462 × 104 |
| IDM | 0.0001 × 104 | 0.0001× 104 | 0.0000 × 104 |
| Sum average | 0.0344 × 104 | 0.0376 × 104 | 0.0364 × 104 |
| Sum variance | 1.7311 × 104 | 1.4712 × 104 | 1.4935 × 104 |
| Sum entropy | 0.0076 × 104 | 0.0082 × 104 | 0.0070 × 104 |
| Difference average | 0.0010 × 104 | 0.0014 × 104 | 0.0013 × 104 |
| Difference variance | 0.022 × 104 | 0.0316 × 104 | 0.0274 × 104 |
Table 3: Shape based features of three tumor slices from the database.
| Slice no | Area (mm2) | Perimeter (mm) |
|---|---|---|
| 1 | 53.252 | 56.65 |
| 2 | 627.783 | 62.15 |
| 3 | 1426.213 | 70.12 |
Table 4: Specification of proposed PSO routine.
| Classifier | Training Efficiency (%) | Testing Efficiency (%) | Hidden neurons | Accuracy (%) | Time (sec) |
|---|---|---|---|---|---|
| ELM with Modified PSO FS | 99.6 | 98.5 | 43 | 99.1 | 45 |
| ELM W/O Modified PSO FS | 97.8 | 96.4 | 57 | 97.8 | 70 |
Table 5: ELM classifier assessment with and without Modified PSO FS.
Conclusion
The major issue in the development of automatic brain tumor tissue classification is the formation of feature extraction analogy with the suitable classifier to attain good accuracy. In this work, an extreme learning machine classifier is suggested for the brain tumor tissue classification along with the modified PSO as feature selector. Simulation result shows that the proposed fusion method of feature extraction works well and produces higher classification accuracy of 99.1% when modified PSO feature extraction methods is applied to ELM. The fused GSIFT and global features along with modified PSO selector shows highest improvement in comparison with other literature studies in neural networks.
References
- Josephine SV, Latha P. Prominence of reducing mortality rate: an anthropological study in the rural of South India. Asian J Res SocSci Human 2016; 6: 929-934.
- Kumar V, Giri VK, Saxena SC. Telemedicine in context to Indian health scenario. J InstEng 2006; 87: 38-45.
- Rohit M, Kabade S, Gaikwad MS. Segmentation of brain tumour and its area calculation in brain MRI images using K-mean clustering and Fuzzy C-mean algorithm, Int J Comp SciEngTechnol 2013; 4: 524-531.
- Yan-shuang H, Yi-xin Y, Jin-hui L. Road vehicle classification based on extreme learning machine. 3rd IntConf Electric Electr 2013; 430-433.
- http://www.ucdenver.edu/academics/colleges/medicalschool/departments/Neurosurgery/patientcare/multi
- JianW,Zhiming C, Sheng VS , Pengpeng Z, Dongliang S, Shengrong G. A comparative study of SIFT and its variants. MeasurSci Rev 2013; 13: 122-131.
- Gladis PRVP, Palani S.A novel approach for feature extraction and selection on mri images for brain tumor classification. Int J Comp Sci Inform Technol 2012; 2: 225-234.
- Kennedy J, Eberhart RC. Particle swarm optimization. Proc IEEE IntConfNeurNetwPiscataw NJ 1995; 1942-1948.
- Tinne T, Krystian M. Local invariant feature detectors: A Survey. Found Trend Comp Graph Vis 2007; 3: 177-280.
- Xu JT, Zhou HM, Huang GB. Extreme learning machine based fast object recognition. IntConf Inform Fus 2012; 1490-1496.