Research Article - Biomedical Research (2018) Volume 29, Issue 8
Effective enrichment of urinary exosomes by polyethylene glycol for RNA detection
Chun-yan Lv* and Tian-lin Yang
Clinical Medical College, Chengdu University of TCM, Chengdu, Sichuan, PR China
- *Corresponding Author:
- Chun-yan Lv
School of Clinical Medicine
Chengdu University of Traditional Chinese Medicine
PR China
Accepted date: February 17, 2018
DOI: 10.4066/biomedicalresearch.29-17-1695
Visit for more related articles at Biomedical ResearchAbstract
Urinary exosome is a kind of extracellular vesicles secreted by the cells of kidney and urinary tract. It is an ideal non-invasive biomarker for obtaining information about the state of renal cells. There are few exosomes in urine which are affected by many factors. Enriching exosomes in urine with inexpensive and efficient method is a major challenge. Because exosomes are virus-sized minuscule membrane vesicles or sacs and people used to enrich virus by Polyethylene Glycol (PEG), here we use PEG to enrich the exosomes in urine of 17 volunteers. To confirm the harvest of exosomes, the morphology of exosomes were observed by Transmission Electron Microscopy (TEM), the common exosomal protein markers (CD63, CD9, tumor susceptibility gene 101, Flotillin-1 and β-actin) were detected by Western blot, and the size distribution was analysed by Malvin Nano-ZS. Next, the exosomal housekeeping gene of miRNA (SnRNA-U6) and mRNA (β-actin) were tested by qRT-PCR to prove that this method could be used in subsequent experiments.
Keywords
Effective enrichment, Urinary exosomes, Polyethylene glycol, RNA detection
Introduction
Exosomes are extracellular vesicles in a proposed diameter of 30-130 nm, similar to virus in size [1-4] that contain various molecules including proteins, lipids and multiple nucleic acids, such as messenger RNA (mRNA) and micro RNA (miRNAs) [5,6], which can be delivered to another cell, and can be functional in the new location [7-9]. Exosomes contain molecular signatures of their progenitor cells and are present in diverse biological fluids including urine [10-12]. Urinary exosomes are mostly from urogenital tract and kidney, they are noninvasive and readily available vesicles and become one of the new fields of biomarker research [13]. Urine is the waste excretion liquid, and liable to be contaminated, easy to be affected by internal and external environment, thus, an inexpensive, efficient and less time-consuming method to enrich exosomes in urine become a major challenge. Currently, the growing attention paid to exploring newer techniques and standardized protocols has been developed, standardized methods or protocols are still lacking. Differential centrifugation is the most commonly used urinary exosomal enrichment procedure [14-16], when combined with sucrose density gradients, exosomes with high puritycan be produced [17]. However, specialized equipment and lengthy protocols are indispensable, its application in the clinical setting is restricted. A variety of commercial precipitation reagents like ExoQuickTM from Systems Biosciences or Total Exosome IsolationTM from Life Technologies are used [18,19]. They are suitable for most laboratories and easy to be utility, however, they are costly, especially if a large number of biological samples need to be processed. Besides that, various techniques including filtration [20-22], microfluidic-based method [23,24], immuno-affinity and peptide-based isolation [25], size-exclusion chromatography [26-28], and so on, have also been proposed for enrichment of exosomes. They have advantages and disadvantages, can be selected according to the experimental command. Polyethylene Glycol (PEG) is a kind of precipitating reagents, which is used to harvest virus particles. Because of the similar dimension of exosomes, some researchers use PEG to enrich successfully exosomes in cell culture medium [29,30]. The urine is a complex mix solution, and easy to be contaminated. We suppose appropriate PEG can also enrich exosomes in urine. Hereupon we present an effective and fast approach for enrich exosomes from urinary supernatant by simply adding PEG solution. Dozens or hundreds of exosomes could be gathered and easily precipitated by low-speed centrifugation. Finally, we evaluated the practical utility of this method by testing housekeeping gene of mRNA (β-actin) and miRNA (snRNA-U6). Overall, we explored a rapid, inexpensive, and adaptable way gathering amounts of exosomes to searching for its role for RNA detection.
Materials and Methods
Research subjects
All the studies were approved by the ethics committee of the Affiliated Hospital of Chengdu University of Traditional Chinese Medicine in accordance with relevant guidelines and regulations. All the volunteers were informed of the experimental study and signed the informed consent form. Seventeen healthy volunteers were enrolled in the study. They were between 22 to 74 (average 55 ± 16.00 y old). Urine routine tests were collected and the results showed: PH value 5.0-7.0 (average 6.33 ± 0.29), conductivity 5.6-29.1 (average 15.1 ± 3.30 ms/cm), urine specific gravity 1.010-1.030 (average 1.013 ± 0.003, Table 1).
| Age | Sex | PH value | Specific gravity | Red cells (/μl) | White cells (/μl) | Epithelial cells (/μl) | Bacteria (/μl) | Conductivity (ms/cm) | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 53 | Male | 6 | 1.015 | 3.8 | 1.9 | 1.6 | 6.5 | 15.3 |
| 2 | 68 | Male | 6.5 | 1.015 | 6 | 3.9 | 4.2 | 55.4 | 11.9 |
| 3 | 71 | Female | 6.5 | 1.01 | 4.6 | 8.1 | 3.4 | 195.5 | 14.2 |
| 4 | 65 | Male | 7 | 1.015 | 12.4 | 4 | 2 | 38.8 | 15 |
| 5 | 57 | Female | 6 | 1.02 | 11.4 | 5.8 | 2.5 | 20 | 14.9 |
| 6 | 37 | Female | 6.5 | 1.01 | 13.7 | 1.9 | 5.6 | 76.5 | 10.4 |
| 7 | 54 | Male | 5.5 | 1.025 | 7 | 1.5 | 0.8 | 6.9 | 15.8 |
| 8 | 45 | Female | 5.5 | 1.015 | 4.8 | 1.7 | 0.8 | 5.2 | 14.4 |
| 9 | 56 | Male | 5.5 | 1.025 | 1.9 | 1.2 | 0.8 | 9.5 | 20.8 |
| 10 | 72 | Female | 5 | 1.025 | 15.3 | 1.1 | 2.3 | 3.4 | 17.3 |
| 11 | 24 | Female | 6.5 | 1.01 | 4.2 | 4.4 | 0.7 | 48.6 | 12 |
| 12 | 44 | Male | 6.5 | 1.03 | 10.3 | 1.1 | 2 | 2.7 | 29.1 |
| 13 | 70 | Male | 6 | 1.02 | 5.5 | 4.3 | 2.5 | 34.7 | 16 |
| 14 | 74 | Female | 6.5 | 1.01 | 3.2 | 1.6 | 4 | 2.6 | 15.4 |
| 15 | 63 | Female | 5.5 | 1.005 | 3.4 | 4.9 | 2.1 | 6 | 5.6 |
| 16 | 22 | Male | 6.5 | 1.015 | 8.1 | 2.1 | 1.2 | 85.5 | 21.9 |
| 17 | 60 | Male | 6.5 | 1.01 | 12.2 | 1.1 | 0.6 | 10.4 | 18.5 |
Table 1. Urine parameters of seventeen volunteers.
Sample collection
20 ml of first morning spot urines were collected from 17 healthy volunteers. Urine samples were centrifuged at 3,000X g for 10 min at 4°C to remove cell debris. The supernatant were gathered and protease inhibitor mixture (0.668 ml 100 mM NaN3, 0.1 ml 100 mM PMSF and 20 μl 1mg/ml leupeptin) were added, and the clarified urine were stored at -80°C as the spares.
Exosome isolation and enrichment
Urine samples stored at -80ºC were taken out and unfreezed at room temperature, an extra vorticity for 30 s was executed. Exosomes in urine were precipitated with 1/2 volume of PEG mother liquor (24% PEG+1 M sodium chloride). The mixture was reversed repeatedly for mixing sufficiently and incubated overnight at 4°C followed by centrifugation at 4,000X g for 1 h at 4°C. We reversed the centrifuge tube and thoroughly cleaned the supernatant with caution. The remained sedimentation at the bottom of the tube was stored at -80°C. Exosome pellets were resuspended in Phosphate Buffered Saline (PBS) before use.
Electron microscopy of exosome samples
10 μl of PBS solution containing exosomes was deposited on copper grid, 10 μl of uranyl acetate was dropped to negatively stain exosomes. The samples were observed by Hitachi electron microscope H-600 (Japan) operated at 80 kV. An Olympus Quemesa Camera was used to record images.
Characterization of exosome and Western blotting analysis
The suspended exosomes were mixed with equal volume of 2X SDS-PAGE protein sample buffer. After a full mixture, samples was added with 1X SDS-PAGE protein buffer to 200 μl, vortexed for 15 s, and denatured in full temperature metal bath at 100°C for 10 min.
10 μl of the prepared loading samples were loaded on the 10% SDS-PAGE gel with extra-long pipet. The gel was run for 40 min at 90 V, then for 60 min at 120 V. The gel was dyed by Bio-SafeTM Coomassie G-250 for 30 min and destained by 20% glacial acetic acid on a rocking plate.
Another gel was used for western blotting analysis, in which the exosomal proteins were separated by 10% SDS-PAGE. After the proteins were transferred onto Polyvinylidene Fluoride (PVDF) membranes (Bio-Rad), the PVDF were blocked in Tris-buffered saline containing 5% albumin bovine V, and incubated with the specific antibodies to exosomal antibodies including CD63 (Abcam, ab8219), CD9 (Abcam, ab92726), tumor susceptibility gene 101 (TSG101, Abcam, ab83), Flotillin 1 (Abcam, ab133497), and β-actin (as an internal control, Zen bioscience, Chengdu), then incubated with appropriate anti-HRP conjugate (Thermo), developed a film in the darkroom.
Nanoparticle tracking analysis
Size distribution within the samples was calculated by tracking the Brownian motion of individual nanoparticles using a Nanoparticle tracking analyzer (NTA, DLS, Nano-ZS, Malvern, Ltd., Malvern, UK). Parameters were set according to the instructions of the NTA instrument, and measuring time is 60 s. 30 μl of urinary exosome was diluted with PBS according to the ratio of 1:2500 into 1 ml, the size and distribution of the urinary exosome was detected and analysed to calculate by the NTA software (version 3.0).
qRT-PCR detection of exosomal housekeeping gene: mRNA (β-actin) and miRNA (snRNA-U6)
Total exosomal RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s recommendations. According to the manufacturer’s protocol, cDNA was synthesized using an Omega Bio-TEK M-MLV Strand cDNA Synthsis Kit (Omega Bio-TEK, USA). Primers were designed by Primer Premier V5.0 software (Table 2). To detect the range of expression of the two candidate genes, reactions were performed using 7500 Real-time quantitative PCR system (ABI, USA). Each sample is detected for two times. The reaction procedure was two steps for 60 cycles: the first step, degeneration for 5 s at 95ºC; the second step, annealing and extension for 60 s at 60ºC. All the runs were completed with a melt curve analysis to confirm the specificity of amplification and lack of primer dimers. Ct (fit point method) and Cp (second derivative method) values were determined by the LightCycler 480 software version 1.2 and exported into an MS Excel data sheet (Microsoft) for analysis after background subtraction.
| Gene | Primer 1 | Primer 2 |
|---|---|---|
| β-actin | 5’-AGCGGGAAATCGTGCGTGACA-3’ | 5’-GTGGACGGGAGAGGACTGG-3’ |
| snRNA-U6 | 5’-CTCGCTTCGGCAGCACA-3’ | 5’-AACGCTTCACGAATTTGCGT-3’ |
Table 2. Primers for β-actin and snRNA U6.
Statistical analysis
The experimental data were represented by means ± SD. The grey level of Western blot result was handled with Image Pro- Plus6.0; log10 method was used to deal with the average Ct value of snRNA U6 and β-actin. The data and chart were processed by Graphpad Prism 6.0.
Results
Urinary exosomal precipitate
After precipitated by PEG overnight and centrifugated for 1 h, a small piece of sediment could be seen in the tube, with gingeli size, gray or yellowish, loosely attached to the bottom of one side of the pipe. Severely shaking and reverse centrifugal tube could cause it to fall off.
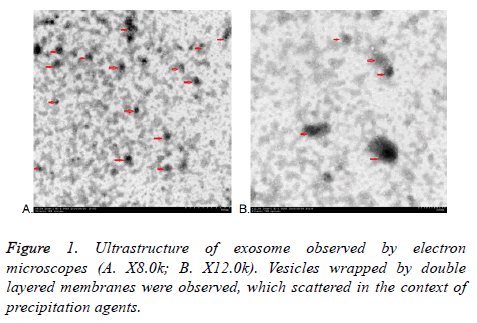
Transmission electron microscope (TEM)
All exosomes are round, which coated with bilayered membrane observed by transmission electron microscopy. They were scattered and less uniform in size with a diameter of about 30-150 nm. Some round particles with typical cupshaped morphology were identified in all samples. This verified that exosomes were harvested. Quantitative analysis of the electron micrographs revealed that the average diameter of vesicle was 75.14 ± 23.3 nm, consistent in the size of exosomes. More or less round materials with high electrons density were observed on the background, their diameters were about 5-15 nm, there were no bilayered membrane structure and these were PEGs (Figure 1).
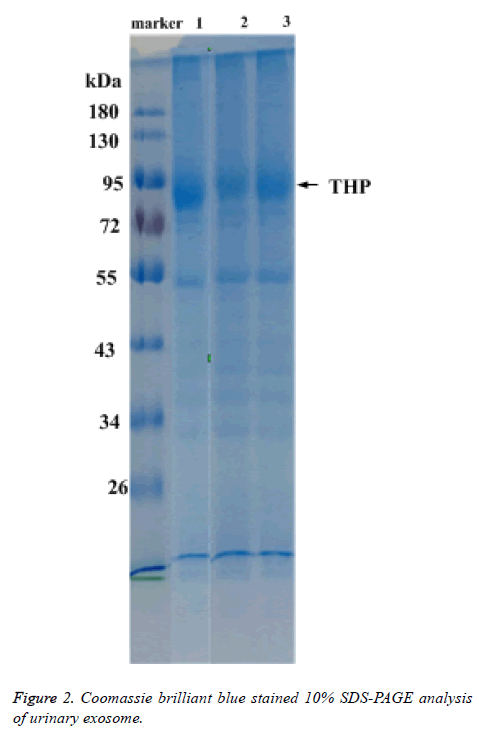
Coomassie brilliant blue stained 10% SDS-PAGE analysis of total proteins
Total protein of urinary exosome with Coomassie brilliant blue staining showed multiple protein bands and covered a large molecular weight range. The bands in the 20-26, 30-43, 50-60 and 72-100 kDa ranges were more visible. A strong protein band of about 95 kDa, putatively identified as Tamn-Horsfall protein, was visible in all exosomal samples. No significant differences were detected between the frequencies of the 17 urinary exosome banding patterns (Figure 2).
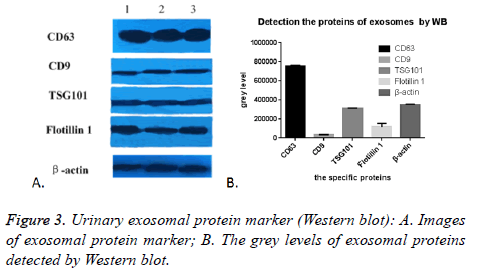
Detection of exosomal protein by Western blot
As expected, we randomly selected five proteins to confirm their presence in urinary exosomes, CD63, CD9, TSG101, Flotillin 1 and β-actin were all successfully detected in all of the 17 cases. The images were shown in Figure 3A. The grey level analyses by Image Pro-Plus 6.0 showed that the expression of CD63 was highly enriched, while the expression of CD9 was the lowest. TSG101 was similar to β-actin, (Figure 3B). This might be due to many factors: the difference exosomes in the urine carrying these five markers, the effect of PEG on these five proteins was not the same. In conclusion, this PEG based approach could enrich and isolate exosomes despite the interference of urinary proteins.
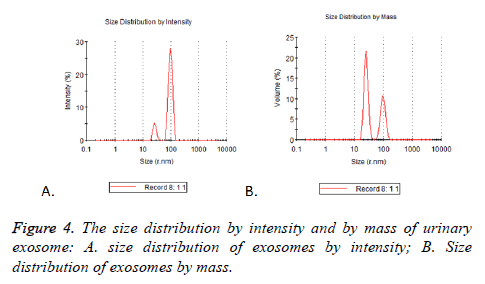
Detection of particle size distribution by nanoparticle tracking analysis (NTA)
Particle mode diameters were found to range between 30 and 130 nm. No significant difference in the size profiles between the 17 cases was observed. Two peaks of size distribution no matter by intensity or by mass were detected by NTA. They were located in 25.37 nm (Figure 4A) and 95.07 nm (Figure 4B), there were 5.2% and 27.9% observed by intensity and 21.5% and 10.6% by mass (Table 3).
| Mode ± SD (nm) | Pd (%) | Est.MW (Kda) (Mode ± SD) | Intensity (%) | Mass (%) | Peak polydispersity | |
|---|---|---|---|---|---|---|
| Peak 1 | 25.37 ± 3.456 | 13.1 | 6.50e+3 ± 1.15e+3 | 13 | 62.6 | Monodispersity |
| Peak 2 | 95.07 ± 15.81 | 16.1 | 1.43e+5 ± 2.01e+4 | 87 | 37.4 | Monodispersity |
Table 3. The size distribution of exosome detected by NTA.
PEG-based method for enrichment urinary exosomes could be used for detecting RNA cargo
Housekeeping gene of mRNA (β-actin) and miRNA (snRNAU6) were detected to test if this PEG-based method for enrichment exosomes in urine could be used for detecting RNA cargo. 17 samples produced strong RNA expression, characteristic of exosomal cargo.
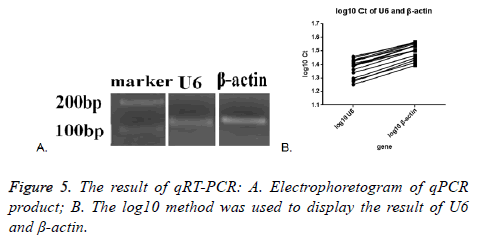
Melt curve and amplification curve of β-actin and snRNA-U6 showed the success of RNA amplification; the result of electrophoretogram of qRT-PCR product showed that the bands of snRNA-U6 and β-actin were about 100 bp, just as we expect (Figure 5A). These data further indicated that vesicles harvested by PEG were not adversely affected by the enrichment protocol, exosomal cargo RNA could therefore be reliably used in downstream analysis.
SnRNA-U6 gene in all the seventeen samples was detected and one in seventeen samples of β-actin was not detected. This might indicate that the expression of small molecules was more stable in the exosomes. The average Ct value was calculated by the two tested results of the same sample, log10 method was used to handle the Ct value. The log10 Ct value of snRNA-U6 was between 1.252-1.459; and that of β-actin was between 1.393-1.565 (Figure 5B). Most of the snRNA-U6 levels were in accord with that of β-actin.
Discussion
Exosomes are membrane-bound vessels that formed by a series of regulatory processes such as endocytosis, fusion and efflux, also known as endocytic multivesicular bodies, a kind of Extracellular Vesicles (EVs). These vesicles can contain cargo both internally and embedded in the membrane, including proteins, lipids, and RNA, despite their small size [31]. It is a hot spot in the field of liquid biopsy markers for the vesicles widely are found in blood, urine, milk and other body fluids [32,33]. Urine is a desirable material for the diagnosis and classification of diseases because of the convenience of its collection in large amounts. Urinary exosome is a kind of extracellular vesicles secreted by the cells of the kidney and urogenital tract. It is potentially a more targeted source of material for biomarker discovery than unfractionated urine, and provides diagnostic and pathophysiological information without an invasive tissue biopsy [34].
But urine is a sort of mixed liquid, the produce and discharge is affected by the physiological and pathological condition, the diet and the medicine [35]. A variety of biological factors in vivo can have an impact on urine volume and component, PH and so on. Therefore, we should pay attention to the process in isolation of exosomes in urine [36,37]. Nowadays, enrichment and analysis of urinary exosomes can be achieved by several methods, although many require specialized equipment or involve lengthy protocols [38-40]. The need for timely analysis and low cost to resolve critical clinical translational bottlenecks has driven considerable innovation with several promising options recently emerging. It is an urgent demand to explore a clinical application method to isolate and enrich urinary exosome which provides an appropriate yield, the lowest possible degree of contamination. Most importance of all, it can be convenient to do in most lab and institution. We have reached the aims by the simple use of PEG, and followed with a low speed centrifuged. Exosomes in urine of 17 healthy volunteers were enriched by this method. The morphological structure of the samples was observed by electron microscopy, the specific marker molecules (CD63, CD9, TSG101 and Flotillin 1) were detected by Western blot and the particle size distribution was analysed by Malvin nanoparticle analyzer, and all of them were in accord with the performance of exosomes. The combination of different approaches proved the harvest of exosomes.
Because of the weak points of low purity, it has always been criticized for isolation of exosomes by precipitation reagents contains volume-excluding polymers (e.g.: PEG, dextrans, or polyvinyls). In this study, the impurity substance of precipitate also can be visible by electron microscope. Moreover, the isolation of urinary exosomes is complicated by the presence of Tamm-Horsfall Protein (THP), which polymerizes and coprecipitates as a contaminant. But exosomes isolated by this method can be used to search the cargo nucleic acid proved by the amplification and detection of exosomal housekeeping gene of miRNA (snRNA-U6) in all 17 cases and most samples of mRNA (β-actin). Because of the cheap price of PEG and other reagents in this process, the non-essential of special equipment such as ultracentrifuge, and simple to operate and be detected in common laboratories and companies; large-scale application especially in low-income countries and regions is expected. Therefore, it is expected that this method is one of the best measurement for the screening of liquid biopsy markers, if the strict detection strategies established, the amount of precipitation agent controlled, and the comparison is appropriate.
Although the β-actin gene in one of the seventeen samples was not detected, this is in accordance with the results obtained from other enrichment methods [41], and therefore this may be related to the RNA content of the larger molecules in exosomes. Then exosmal miRNA maybe the more stable molecular for detection.
In summary, the methodology described in this paper is a wellestablished approach for enrichment exosomes. This approach is quick, cheap and convenient that making feasible the analysis in a clinical facility. This method allows the possibility to study mRNA and miRNA of urinary exosome and it is especially suitable for small or medium research institutes and basic medical units.
Acknowledgments
This work is supported in part by research grants from the fund of Chengdu University of Traditional Chinese Medicine (Project No.CGPY1601).
References
- Anirban G, Michelle D, Ian CC, Steven GG. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS One 2014; 9: 110443.
- Rani S, OBrien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, Crown J, ODriscoll L. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol Biol 2011; 784: 181-195.
- Marina C, Graca R, Clotilde T. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014; 30: 255-289.
- Guoku H, Lu Y, Yu C, Fang N. Emerging roles of extracellular vesicles in neurodegenerative disorders: focus on HIV-associated neurological complications. Cell Death Dis 2016; 7: 2481.
- Valadi H. Exosomes contain a selective number of mRNA and microRNA. Allergy 2007; 62.
- Arsen OB, Igor VK. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3-untranslated regions. Biol Direct 2013; 8: 12.
- Yez MM, Siljander RM, Andreu Z, Zavec AB. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015; 4: 27066.
- Chaput N, Thery C. Exosomes: immune properties and potential clinical implementations. Semin Immunopathol 2011; 33: 419-440.
- Md J, Shair KH, Marquitz AR, Kung CP. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci USA 2010; 107: 20370-20375.
- Miranda KC, Bond DT, McKee M, Skog J. Nucleic acids within urinary exosomes/ microvesicles are potential biomarkers for renal disease. Kidney Int 2010; 78: 191-199.
- Camussi G, Deregibus MC, Bruno S, Grange C. Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res 2011; 1: 98-110.
- Endzelins E, Berger A, Melne V, Bajo-Santos C. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017; 17: 730.
- Huebner AR, Somparn P, Benjachat T, Leelahavanichkul A, Avihingsanon Y, Fenton RA, Pisitkun T. Exosomes in urine biomarker discovery. Adv Exp Med Biol 2015; 845: 43-58.
- Gonzales PA, Zhou H, Pisitkun T, Wang NS, Star RA, Knepper MA, Yuen PS. Isolation and purification of exosomes in urine. Methods Mol Biol 2010; 641: 89-99.
- Szempruch AJ, Sykes SE, Kieft R, Denniso L. Extracellular vesicles from trypanosoma brucei mediate virulence factor transfer and cause host anemia. Cell 2016; 164: 246-257.
- Tataruch-Weinert D, Musante L, Kretz O, Holthofer H. Urinary extracellular vesicles for RNA extraction: optimization of a protocol devoid of prokaryote contamination. J Extracell Vesicles 2016; 5: 30281.
- Schageman J, Zeringer E, Li M, Barta T. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int 2013; 2013: 253957-253957.
- Laulagnier K, Javalet C, Hemming FJ, Sadoul R. Purification and analysis of exosomes released by mature cortical neurons following synaptic activation. Methods Mol Biol 2017; 1545: 129-138.
- Yan IK, Shukla N, Borrelli DA, Patel T. Use of a hollow fiber bioreactor to collect extracellular vesicles from cells in culture. Methods Mol Biol 2018; 1740: 35-41.
- Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, Star RA. Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol 2007; 292: 1657-1661.
- Musante L, Tataruch DE, Holthofer H. Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front Endocrinol (Lausanne) 2014; 5: 149.
- Merchant ML, Powell DW, Wilkey DW, Cummins TD, Deegens JK, Rood IM, McAfee KJ, Fleischer C, Klein E, Klein JB. Microfiltration isolation of human urinary exosomes for characterization by MS. Proteomics Clin Appl 2010; 4: 84-96.
- Contreras-Naranjo JC, Wu HJ, Ugaz VM. Microfluidics for exosome isolation and analysis: enabling liquid biopsy for personalized medicine. Lab Chip 2017; 17: 3558-3577.
- Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014; 14: 1891-900.
- Kang YT, Kim YJ, Bu J, Cho YH. High-purity capture and release of circulating exosomes using an exosome-specific dual-patterned immunofiltration (ExoDIF) device. Nanoscale 2017; 9: 13495-13505.
- Stranska R, Gysbrechts L, Wouters J, Vermeersch P. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J Transl Med 2018; 16: 1.
- Kreimer S, Ivanov AR. Rapid isolation of extracellular vesicles from blood plasma with size-exclusion chromatography followed by mass spectrometry-based proteomic profiling. Methods Mol Biol 2017; 1660: 295-302.
- Lobb R, MOller A. Size exclusion chromatography: a simple and reliable method for exosome purification. Methods Mol Biol 2017; 660: 105-110.
- Rider MA, Hurwitz SN, Dgm J. ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep 2016; 6: 23978.
- Weng Y, Sui Z, Shan Y, Hu Y, Chen Y, Zhang L, Zhang Y. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst 2016; 141: 4640-4646.
- Shen C, Hao SG, Zhao CX, Zhu J, Wang C. Antileukaemia immunity: effect of exosomes against NB4 acute promyelocytic leukaemia cells. J Int Med Res 2011; 39: 740-747.
- Perakis S, Speicher MR. Emerging concepts in liquid biopsies. BMC Med 2017; 15: 75.
- Zhang W, Xia W, Lv Z, Ni C, Xin Y, Yang L. Liquid biopsy for cancer: circulating tumor cells, circulating free DNA or exosomes? Cell Physiol Biochem 2017; 41: 755-768.
- Street JM, Koritzinsky EH, Glispie DM, Star RA, Yuen PS. Urine exosomes: an emerging trove of biomarkers. Adv Clin Chem 2017; 78: 103-122.
- Salih M, Fenton RA, Knipscheer J, Janssen JW, Vredenbregt-van den Berg MS, Jenster G, Zietse R, Hoorn EJ. An immunoassay for urinary extracellular vesicles. Am J Physiol Renal Physiol 2016; 310: 796-801.
- Yao Zhao, Kaiyong Chen, Haining Li, Huiyi Wu. Effect of pH on the isolation of urinary exosome. Int Urol Nephrol 2017; 49: 165-169.
- Puhka M, Nordberg ME, Valkonen S, Rannikko A, Kallioniemi O, Siljander P, Af Hallstrom TM. KeepEX, a simple dilution protocol for improving extracellular vesicle yields from urine. Eur J Pharm Sci 2017; 98: 30-39.
- Mol EA, Goumans MJ, Doevendans PA, Sluijter JP, Vader P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomedicine 2017; 1549-9634: 30054-30060.
- Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, Hamrick MW, Liu Y. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One 2017; 12: 0170628.
- Xu R, Simpson RJ, Greening DW. A protocol for isolation and proteomic characterization of distinct extracellular vesicle subtypes by sequential centrifugal ultrafiltration. Methods Mol Biol 2017; 1545: 91-116.
- Hendriks RJ, Dijkstra S, Jannink SA, Steffens MG. Comparative analysis of prostate cancer specific biomarkers PCA3 and ERG in whole urine, urinary sediments and exosomes. Clin Chem Lab Med 2016; 54: 483-492.