- Biomedical Research (2013) Volume 24, Issue 1
Differential expressions of PDGF-BB and their distributions in PDGF-BB down-regulated transgenic mouse.
Yan-Bin XiYang1, Ya-Zhao1, Bing-Tuan Lu1, Jin Ru1 , Wei Zhang3, Xiong-Zhi Quan3, Lian-Feng Zhang 1 , and Ting-Hua Wang1,2*z
1Institute of Neuroscience, Kunming Medical University, Kunming 650500, China
2Institute of Neurological Disease, West China Hospital, Sichuan University, Chengdu 610041, China
3Key Laboratory of Human Diseases Comparative Medicine, Ministry of Health; Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences(CAMS) &Comparative Medicine Centre, Peking Union Medical College (PUMC), Beijing 100021, China
- *Corresponding Author:
- Ting-Hua Wang
Director, Institute of Neurological Disease
West China Hospital, Sichuan University
Chengdu, China; Director, Institute of Neuroscience
Kunming Medical University, Kunming
China
Accepted date: October 21 2012
Abstract
As a growth regulator, PDGF-BB plays a crucial role in regulating neuronal survival and cell differentiation during embryonic development and in the adulthood. The multiple roles and mechanisms of PDGF-BB are still not known, especially involved in plasticity after spinal cord injury. However, before we can carry out further investigations a transgenic animal model with PDGF-BB down-regulation is needed. In the present study, expressional silencing PDGF-BB was achieved by select predesigning hairpins targeting mouse PDGF-BB genes. Six homozygous transgenic offspring were generated and the protein expressions of PDGF-BB were detected in multiple tissues of these mice. The down-regulated rates of PDGF-BB in different transgenic mice were also evaluated. Results showed that different PDGF-BB expressions were detected in multiple tissues and protein levels of PDGF-BB reduced at different rates by relative to that of wild type ones. The expressions of PDGF-BB proteins in transgenic mice decreased at most by 56%. The present study generated TG mice with PDGF-BB down-regulation and established mice model for systemic exploring the possible roles of PDGF-BB in vivo in different pathology conditions.
Keywords
PDGF-BB, down-regulation, transgenic mouse, expression, distribution
Introduction
Traumatic injury to the spinal cord results in irreversible locomotor, sensory, and autonomic dysfunctions for which there is currently no effective pharmacologic treatment [1]. Following spinal cord injury (SCI), cell death, neuroinflammation, tissue degradation, and reactive astrogliosis profoundly modify the extracellular matrix and foster the generation of an inhibitory environment to neural repair and regeneration [2,3].
The platelet-derived growth factor (PDGF) family contains five members found naturally in the body. These are AA homodimer, AB heterodimer, BB homodimer, CC homodimer, and DD homodimer. PDGF-BB is a homodimer of two antiparallel B-chains covalently linked through two disulfide bonds [4,5]. Two receptor subtypes of PDGF (PDGFR-α and PDGFR-β) can form mature dimeric receptor complexes that can bind to ligands with different affinities [4]. PDGFR-α is largely expressed in oligodendroglia progenitors, while PDGFR-β is predominantly expressed in neurons [5] and upregulated in the neonatal rat brain [6]. PDGF-BB contributes to tissue repair through its multiply biological activities. It is reported that PDGF-BB: i) is a primary component of platelets released at sites of injury following platelet activation; ii) is chemotactic and mitogenic for cells of mesenchymal origin [7,8]; iii) is proangiogenic, upregulating vascular endothelial growth factor (VEGF) to stimulate new capillary growth [7]. Moreover, PDGF-BB that specifically binds to PDGFR-ββ is abundantly expressed in neurons and is upregulated in neonatal brains [9,10]. However, the possible roles of PDGF-BB in neuroplasticity after spinal cord injury (SCI) were still unknown and study of associated signaling mechanisms has been limited by the lack of a robust animal model.
In the present study, in order to explore the roles of PDGF-BB in neuroplasticity after SCI, we established transgenic (TG) mice with PDGF-BB gene downregulation (PDGF-BB-do). Polymers chain reaction (PCR) were performed to identify the genotypes of mice.
Then, Western blot and immunohistochemistry (IHC) were employed to detect the protein expressional levels and distributions of PDGF-BB in multiply tissues of different genotypes TG mice, including olfactory bulb, cortex, frontal lobe, basal forebrain, cerebellum, hypothalamus, medulla oblongata, spinal cord, trachea, lung, heart, liver, spleen, kidney, adrenal gland, intestines, skeletal muscles and epidermis. The rates of PDGF-BB down-regulation in multiple tissues of different genotypes were evaluated by relative intensity to the level of wild type (WT). The present study generated TG mice with PDGF-BB down-regulation for the first time and supplied systemic distributions of PDGF-BB protein in TG mice, therefore provided a critical tool for exploring roles of PDGF-BB in different disease conditions in vivo.
Materials and Methods
Animal generation
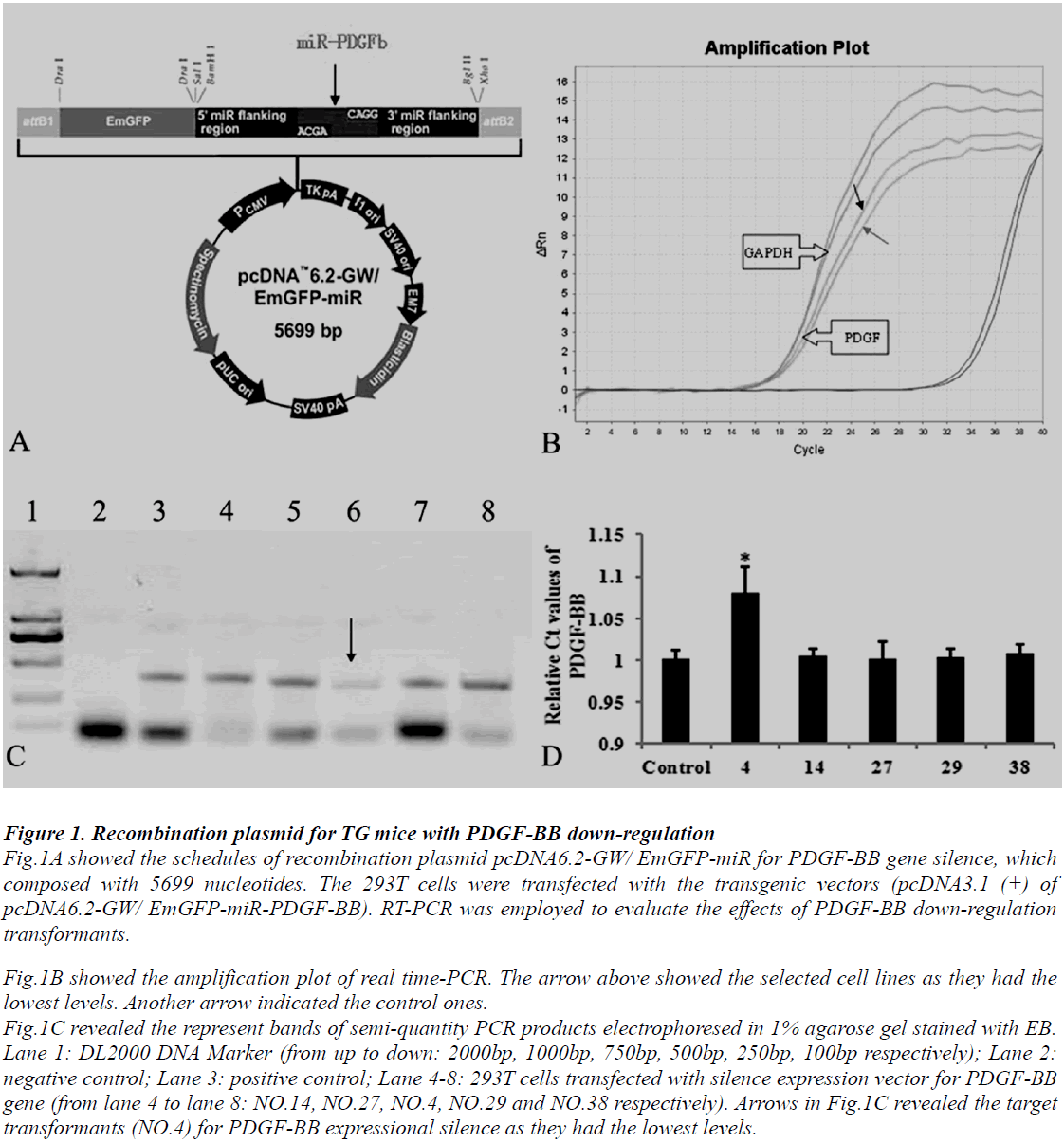
Animal use and care were in accordance with the animal care guidelines, which conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996). PDGF-BB transgenic mice with C57BL/6J genetic background were produced by our collaborators in The Institute of Laboratory Animal Science (Chinese Academy of Medical Sciences & Comparative Medicine Centre, Peking Union Medical College, Beijing, China). The protocols are as follows: at least four silence expression sites of PDGF-BB were designed by software supplied by Invitrogen Company, USA. Then select predesigned short hairpins RNA (shRNAs) targeting mouse PDGF-BB gene (Mus musculus, GeneID: 18591) and the reconstruction plasmid were designed (Fig.1A) and purchased also from Invitrogen Company. The constructed recombinant plasmid was transferred into 293T cells. The transformants were screened and identified by polymers chain reaction (PCR) detections and restriction analysis (Fig.1B, C and D). The protocol of PCR was described following. The transgene was then isolated from the cloning plasmid and purified by Avr II digestion, followed by diluted to a final concentration of 5 ng/μL. The final transgenic fragment was microinjected into fertilized mouse eggs (F1 [C57BL/6 x CBA/J] x F1 [C57BL/6 x CBA/J]).
Figure 1: Recombination plasmid for TG mice with PDGF-BB down-regulation Fig.1A showed the schedules of recombination plasmid pcDNA6.2-GW/ EmGFP-miR for PDGF-BB gene silence, which composed with 5699 nucleotides. The 293T cells were transfected with the transgenic vectors (pcDNA3.1 (+) of pcDNA6.2-GW/ EmGFP-miR-PDGF-BB). RT-PCR was employed to evaluate the effects of PDGF-BB down-regulation transformants. Fig.1B showed the amplification plot of real time-PCR. The arrow above showed the selected cell lines as they had the lowest levels. Another arrow indicated the control ones. Fig.1C revealed the represent bands of semi-quantity PCR products electrophoresed in 1% agarose gel stained with EB. Lane 1: DL2000 DNA Marker (from up to down: 2000bp, 1000bp, 750bp, 500bp, 250bp, 100bp respectively); Lane 2: negative control; Lane 3: positive control; Lane 4-8: 293T cells transfected with silence expression vector for PDGF-BB gene (from lane 4 to lane 8: NO.14, NO.27, NO.4, NO.29 and NO.38 respectively). Arrows in Fig.1C revealed the target transformants (NO.4) for PDGF-BB expressional silence as they had the lowest levels.
Transgenic mice were mated with nontransgenic partners to maintain heterozygosity of the transgene or with transgenic partners to generate homozygous transgenic offspring. In the latter case, transgenic male mice were test mated with two wild-type female mice, and the offspring (15-20 individuals) was analyzed by PCR. Male mice that produced exclusively transgenic offspring were considered homozygous for the transgene. PDGF-BB TG mice and their age-matched, non-transgenic littermates (WT mice) were used.
Real-time PCR
Total RNA was isolated from the harvested cells by using Trizol reagent (Gibco Life Technologies, Rockville, MD). cDNA was synthesized by using Oligo(dT)18 and MMLV reverse transcriptase (Promega, Madison, WI). Primers employed were synthesized by Shengon (Shanghai, China) and were described as follows. For detections in expression of PDGF-BB mRNA, the following primers were used: sense, 5' CACTCCATCCGCTCCTTT 3'; antisense, 5' CTTCTTTCGCACAATCTCAAT 3'. For GAPDH detections, the following primers were used: sense, 5' CAAGGTCATCCATGACAACTTTG 3'; anti-sense, 5' GTCCACCACCCTGTTGCTGTAG 3'. The cDNA was 10-fold serially diluted to seven concentrations for the standard curve.
To analyze the mRNA expressions of PDGF-BB in transformants, RT-PCR protocol was applied using an ABI 5700 instrument (Bio-Rad). Reactions were performed in a 20 μl volume with 0.25 μM primers, 5 mM MgCl2, nucleotides, Taq DNA polymerase, and buffers were included in the DNA Master SYBR Green I mix (Applied Biosystems). Specificity of amplification products was confirmed by melting curve analysis. PCR was performed by the denaturation step at 95°C for 3 minutes, followed by 35 cycles of 95°C for 10 seconds, 58°C for 10 seconds, and 72°C for 30 seconds. Fluorescent signals from PCR products were recorded at 85.5°C for 5 seconds. PDGF-BB mRNA levels were normalized as the ratio of the fluorescence intensity from PDGF-BB to that of GAPDH.
Semi-quantity PCR
Preparations for RNA samples were as described above. Then the total RNA was eluted in 20μl RNase-free Water (Gibco Life Technologies, Rockville, MD). The RNA was kept on ice and their concentrations were measured by a Nanodrop spectrophotometer (ND-1000). An equal amount of RNA (4μg) was used for each experiment. Semi-quantity PCR for the PDGF-BB was performed. The following primers were used: sense, 5' CACTCCATCCGCTCCTTT 3'; anti-sense, 5' CTTCTTTCGCACAATCTCAAT 3'. The product length of PCR is 381bp. Gene primers were synthesized by TaKaRa Company.
Experiments were duplicated to verify the results. For RNA amplification, the first-strand cDNA was synthesized from 4μg of total RNA, using Revert AidTM First Strand cDNA Synthesis Kit (Fermentas Company, U.S.A.). PCR was then carried out using the PCR Master Mix Kit (Fermentas Company, U.S.A.) for 35 cycles, consisting of denaturation at 94°C for 1 min, annealing for 1 min, and extension at 72°C for 1 min. Then RT-PCR products were electrophoresed in 1% agarose gel stained with ethidium bromide (EB) and visualized, using an ultra violet gel imager (BIO-GEL, BIP-RAD). The image analysis was performed by SYN Gene Tool (LIVE Science, U.S.A.).
Assessment of genotypes
The inserted fragment was identified by PCR. For PDGFBB down-regulated (PDGF-BB-do) lines, the following primers were used: sense, 5′GAGCAAAGACCCCAACGAG 3′; antisense, 5′TTATGAACAAACGACCCAACAC3′. The product length of PCR is 419bp. Briefly, PCR was carried out using the PCR Master Mix Kit (Fermentas Company, U.S.A.) for 35 cycles, consisting of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. Then RTPCR products were electrophoresed in 1% agarose gel stained with EB and visualized, using an ultra violet gel imager (BIO-GEL, BIP-RAD). The image analysis was performed by SYN Gene Tool (LIVE Science, U.S.A.).
Expressions of PDGF-BB Protein in different TG mouse
To detect the level of PDGF-BB protein, multiply tissues described as above were obtained from mice with different genic genotypes and WT ones. After carefully rinsing in cooled PBS, the tissues were homogenized on ice in a Lysis Buffer (Beyotime, P0012, China), and centrifuged at 12,000rp for 30 min. The supernatant was then obtained and stored at -80°C for later use. Protein concentration was assayed with BCA reagent (Sigma, St. Louis, MO, USA). A 20 μl aliquot of the samples was loaded on to each lane and electrophoresed on 12% SDSpolyacrylamide gel (SDS-PAGE) for 2.5 h at a constant voltage of 120 V. Proteins were transferred from the gel to a nitrocellulose membrane for 6.5h at 24 V. The membrane was blocked with phosphate-buffered saline containing 0.05% Tween-20 (PBST) with 10% nonfat dry milk overnight at 4°C for 12h, then the membrane was washed three times for 10min each time. They were then rinsed with PBST and incubated with the primary antibody for PDGF-BB (1:800, Chemican) at 4°C for 24h. After washing, the membrane was incubated with a HRPconjugated goat anti-rabbit IgG (1:5,000; Vector Laboratories, CA) for 2h at room temperature, and washing as described above. The membrane was developed in ECM kit, and then pictured by Bio-Gel Imagining system equipped with Genius synaptic gene tool software. Densitometry analysis for PDGF-BB protein was performed. Βeta-tubulin (1:500, Santa cruz) was used as internal control.
Immunohistochemistry
The tissue preparation and procedure of immunohistochemistry used in this study has been introduced previously [11]. Briefly, multiply tissues from TG mice described above were harvested and postfixed. Sections of 20μm thickness were cut in a freezing microtome (Leica CM1900, Germany), collected in a plate of 24 wells and rinsed. After immersing in 0.01M PBS containing 5% goat non-immune serum and 0.3% TritonX-100 solution at 37°C for 30min, they were subsequently incubated for 48h at 4°C with 2% goat serum containing goat polyclonal antibodies PDGF-BB (1:800, Santa Cruz).
They were washed and incubated in Reagents I and II from the Reagent Kit (Chemicon, Anti-Rabbit/Mouse Poly-HRP IHC Detection Kit, USA), each for 30min at 37°C. It was again rinsed five times, each for 5min in 0.01M PBST. Finally, sections were detected by DAB staining. Negative control was performed by replacing the primary antibody with 2% goat serum to ascertain the specificity of antibody staining. Immunoreactive products were observed and photographed with a light microscope (Leica. DMIRB, Germany) coupled with a computer assisted video camera.
Statistical analysis
All data were expressed as the mean ± S.E.M. They were analyzed using One-way ANOVA and LSD-q test by SPSS software package. Statistical significance was defined as P<0.05.
Results
Genotypes detection of TG
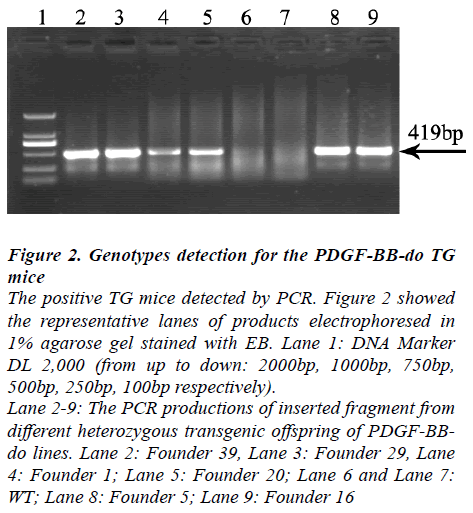
Six heterozygozity transgenic offspring of PDGF-BB-do TG lines were obtained. All of them could produce offspring, which were designated Founder 39, Founder 29, Founder 1, Founder 20, Founder 5 and Founder 16. The TG mice with inserted fragment, identified by PCR, were regarded as positive TG
Figure 2: Genotypes detection for the PDGF-BB-do TG mice The positive TG mice detected by PCR. Figure 2 showed the representative lanes of products electrophoresed in 1% agarose gel stained with EB. Lane 1: DNA Marker DL 2,000 (from up to down: 2000bp, 1000bp, 750bp, 500bp, 250bp, 100bp respectively). Lane 2-9: The PCR productions of inserted fragment from different heterozygous transgenic offspring of PDGF-BBdo lines. Lane 2: Founder 39, Lane 3: Founder 29, Lane 4: Founder 1; Lane 5: Founder 20; Lane 6 and Lane 7: WT; Lane 8: Founder 5; Lane 9: Founder 16
Protein expressional changes of PDGF-BB in multiple tissues of TG with different genotype
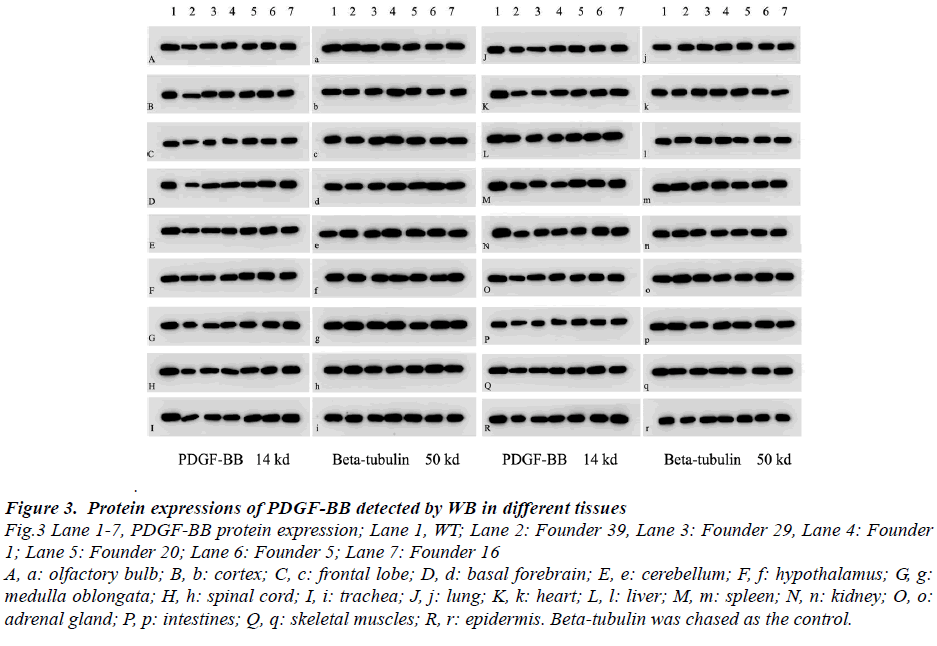
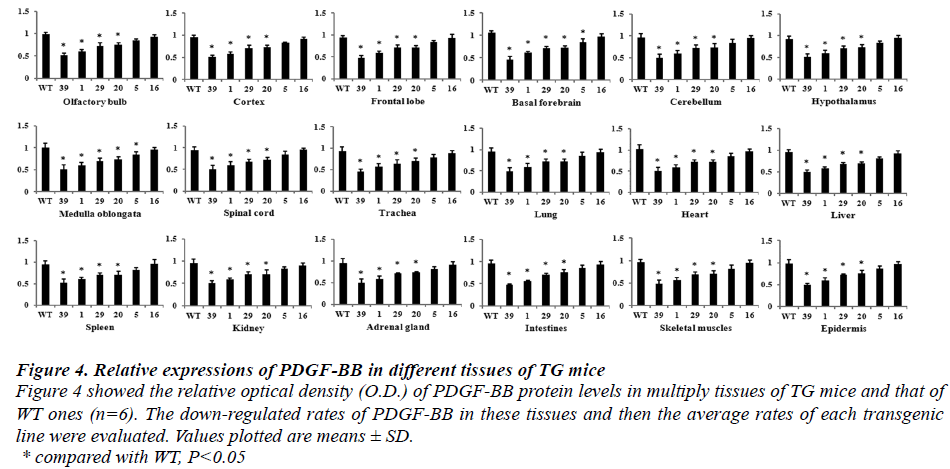
Results of Western blot, which detected in different multiple tissues of six genotypes TG (Founder 39, Founder 29, Founder 1, Founder 20, Founder 5 and Founder 16), indicated that PDGF-BB expressions were down-regulated by different percent in these TG mice (Fig.3 and Fig.4). The rates of protein down-regulated calculated as following, rates of protein down-regulation = O.D. (WT - Founder)/ O.D.WT *100% (O.D. means optical density). The results revealed that all of the TG mice, but Founder 5 and Founder 16, showed significant difference compared with that of the WT one
Figure 3: Protein expressions of PDGF-BB detected by WB in different tissues Fig.3 Lane 1-7, PDGF-BB protein expression; Lane 1, WT; Lane 2: Founder 39, Lane 3: Founder 29, Lane 4: Founder 1; Lane 5: Founder 20; Lane 6: Founder 5; Lane 7: Founder 16 A, a: olfactory bulb; B, b: cortex; C, c: frontal lobe; D, d: basal forebrain; E, e: cerebellum; F, f: hypothalamus; G, g: medulla oblongata; H, h: spinal cord; I, i: trachea; J, j: lung; K, k: heart; L, l: liver; M, m: spleen; N, n: kidney; O, o: adrenal gland; P, p: intestines; Q, q: skeletal muscles; R, r: epidermis. Beta-tubulin was chased as the control.
Figure 4: Relative expressions of PDGF-BB in different tissues of TG mice Figure 4 showed the relative optical density (O.D.) of PDGF-BB protein levels in multiply tissues of TG mice and that of WT ones (n=6). The down-regulated rates of PDGF-BB in these tissues and then the average rates of each transgenic line were evaluated. Values plotted are means ± SD. * compared with WT, P < 0.05 According to formula of the down-regulated rates of PDGF-BB protein, the average down-regulated rates of the six transgenic lines were calculated and described as followed. The average down-regulated rates were 56%, 48%, 22%, 20%, 12% and 2% in Founder 39, Founder 1, Founder 29, Founder 20, Founder 5 and Founder 16, respectively. The relative expressions of PDGF-BB proteins of Founder 39, Founder 1, Founder 29 and Founder 20 showed significant different compared with that of WT ones (P<0.05).
Distributions of PDGF-BB in multiple tissues
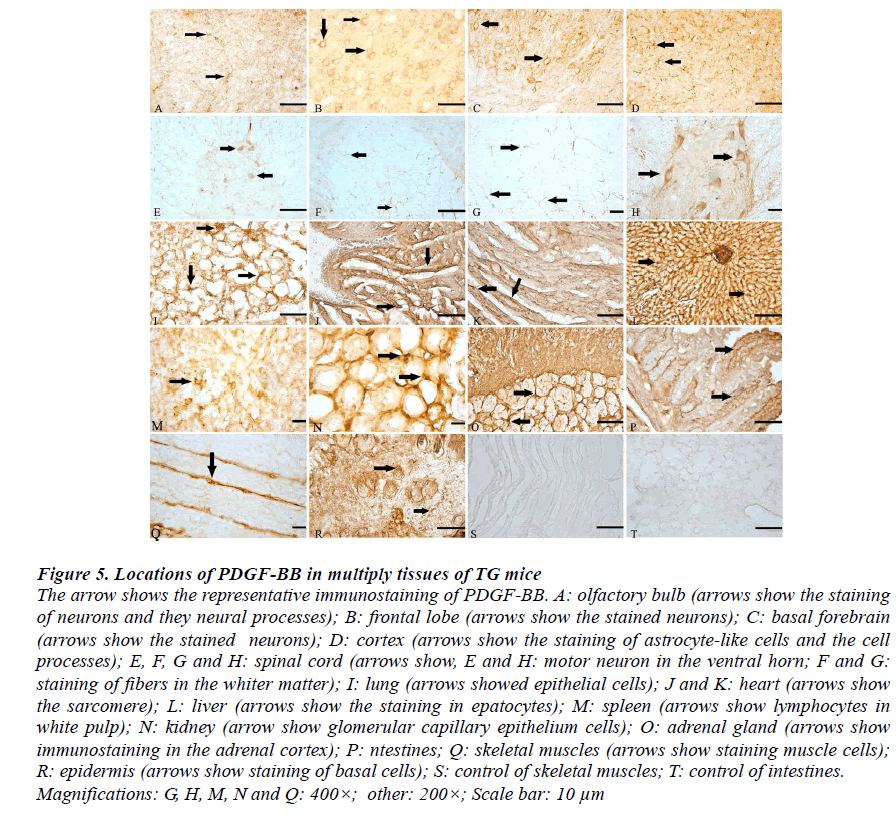
Control of immunostaining specificity was performed by replacing the primary antibody with 2% goat serum. These controls did not exhibit any specific immune-staining in the epidermis and intestines (Fig.5S and T, respectively). The immunoreactions (IR) of PDGF-BB could be seen in the cytoplasm and processes, but not in the nucleus.
Olfactory bulb
IR of PDGF-BB was seen in basal cells, supporting cells, neurons, apical cytoplasmic region of olfactory epithelium, lamina propria and gland's cell cytoplasm (Fig.5H).
Brain
The distributions of PDGF-BB immunopositive neurons and glia-liked cells were observed within the cortex, basal brain, frontal lobe, cerebellum, hypothalamus, medulla oblongata. They occurred in all layers of the cortical regions examined in this study, including I external pyramidal layer and internal pyramidal layer. Morphologically, they were mainly pyramidal neurons, and most were medium- and small-sized cells. The somata and proximal dendrites with PDGF-BB IR were also observed in different subpopulations of neurons in the brain stem (Fig. 5A-D).
Spinal cord
The PDGF-BB immunopositive profiles were present in rostral horn, ventral horn neurons as well as white matter of the spinal cord (Fig.5E, F and G).
Lung
The immunostaining of PDGF-BB was found seen in the epithelial cells, vascular endothelial cell, as well as white blood cell (Fig.5I).
Liver
PDGF-BB was distributed in the cytoplasm of hepatocytes throughout the liver lobule. IR of PDGF-BB was also seen in liver acinus, sinusoid and vascular endothelial cell (Fig.5L).
Spleen
IR of PDGF-BB was detected in Tunica media of artery, subendothelial smooth muscle cell and endothelia cell (Fig.5M).
Kidney
Representative IR for PDGF-BB in renal section of TG mice showed diffuse positive staining within renal cortex, medullary interstitial cells, as well as the peritubular capillaries (Fig. 5 N).
Adrenal gland
The majority of PDGF-BB positive cells are located directly underneath the capsule, in the adrenal cortex (Fig.5O).
Intestine
PDGF-BB immunopositive files dispersed in lamina propria, epithelium mucosae and muscular layer. The immune-positive staining was primarily in the cytoplasm and partial cytolemma (Fig.5P).
Muscle
In agreement with the known localization, PDGF-BB staining was localized to the sarcolemma in skeletal muscle of mice (Fig.5Q). In the sarcoplasm there was staining in a transverse striation pattern at regular intervals the length of a sarcomere. Immunostaining for PDGF-BB also showed positive staining in coronary arteries of hearts (Fig.5J and K).
Epidermis
The positive-reactions of PDGF-BB were detected in the epidermis of TG mice. The IR was found in cytoplasm and cytolemma of basal cells and follicular epithelium (Fig.5R).
Figure 5: Locations of PDGF-BB in multiply tissues of TG mice The arrow shows the representative immunostaining of PDGF-BB. A: olfactory bulb (arrows show the staining of neurons and they neural processes); B: frontal lobe (arrows show the stained neurons); C: basal forebrain (arrows show the stained neurons); D: cortex (arrows show the staining of astrocyte-like cells and the cell processes); E, F, G and H: spinal cord (arrows show, E and H: motor neuron in the ventral horn; F and G: staining of fibers in the whiter matter); I: lung (arrows showed epithelial cells); J and K: heart (arrows show the sarcomere); L: liver (arrows show the staining in epatocytes); M: spleen (arrows show lymphocytes in white pulp); N: kidney (arrow show glomerular capillary epithelium cells); O: adrenal gland (arrows show immunostaining in the adrenal cortex); P: ntestines; Q: skeletal muscles (arrows show staining muscle cells); R: epidermis (arrows show staining of basal cells); S: control of skeletal muscles; T: control of intestines. Magnifications: G, H, M, N and Q: 400×; other: 200×; Scale bar: 10 μm
Discussion
The present study is the first one to generate different expressional level of PDGF-BB transgenic mice, which displayed that delivering with interference shRNAs special to PDGF-BB gene induced PDGF-BB protein expressional decrease in TG mice, especially in central nervous system. Also, the expressional decrease in PDGFBB protein showed diverse in different phenotypic transgenic lines. The results detected by Western blot showed that the relative highest value (nearly to 56%) was detected in Founder 39, while the lowest was detected in Founder 16 (2%). In addition, we explored the systemic distribution of PDGF-BB in different tissues of TG mice. The present study created serials new TG mice models of PDGF-BB down-regulation and reported the intimate and systematic distributions of PDGF-BB in TG mice for further investigations.
1. The establishing of PDGF-BB down-regulated TG mice model
Our results of PCR for genotypes detection, which showed that the inserted fragments (419bp) were detected in the six TG offspring of PDGF-BB-do lines, indicated that new TG mice model of PDGF-BB-do lines were obtained successfully by genetic manipulation. This study generated six kinds of available TG mice, which were designated Founder 39, Founder 29, Founder 1, Founder 20, Founder 5 and Founder 16. Hairpin RNAs, consisting of long duplex structures, have been proved as effective triggers of stable gene silencing in both of plants and animals. As short hairpin precursors, processed into active, 27-nt RNAs by Dicer, and recognize target mRNAs via base-pairing interactions, thus permitting the construction of transgenic animals in which RNAi enforces stable and heritable gene silencing [12, 13]. The present results showed that delivering transformer consisted with shRNAs for PDGF-BB induced extensively PDGF-BB down-regulation in mice. Random integration of transgenic fragment effectively reduced the expressions of PDGF-BB in TG mice, especially in center nerves system. However, the results revealed that diverse down-regulated rates of PDGF-BB protein were detected in different phenotypic lines. The diverse expressional changes of PDGF-BB protein in six kinds of TG mice might be resulted from the randomly inserted sites of the recombination vectors in the target gene. Furthermore, some unknown mechanisms of posttranscription regulation in different tissues might induce the different suppression in PDGF-BB expressions of multiple tissues [14].
2. The known roles of PDGF-BB and the discovery of our study
As a multiple promoter factor, PDGF-BB is wide-spread in all organs and tissues. PDGFs are growth-regulatory molecules that stimulate chemotaxis, proliferation, and increased metabolism of primarily connective tissue cells [9]. PDGF-BB is widely expressed in both neurons and glial cells throughout the CNS [15]. After combining with PDGF-R-beta, PDGF-BB can induce a series of biological effects such as DNA synthesis, cell proliferation and differentiation, cytokines chemotaxis and apoptosis [16, 17]. PDGF-BB also promotes mitosis of glial cells and chemotaxis for mesenchyme-derived cells [17, 18]. Therefore, PDGF-BB plays important roles not only under normal physiological but also pathophysiological conditions in the central nervous system (CNS).
Reports had demonstrated that immunostaining for PDGF BB-chain was located in neurons, principal dendrites, some axons, and probable terminals throughout the brain and the spinal cord of mice, which was similar with the finds of the present study [15]. However, there were a few reports about the systemic distributions of PDGF-BB, especially in mice [15, 18]. The present study revealed the intimate and systematic distributions of PDGF-BB protein of PDGF-BB-do TG mice. These results indicated that PDGF-BB might play multiply biologic roles according to the different cell type. Furthermore, the present work indicated that the transgenic manipulation made no effects on the distributions of PDGF-BB in TG mice.
Conclusion
In conclusion, the present study established new transgenic mice lines with extensive PDGF-BB downregulation. We also supplied the down-regulated rates and systemic distributions of PDGF-BB protein in six phenotypic transgenic mice. The results showed that Founder 39, Founder 1, Founder 29 and Founder 20 might be designed as the target lines for further researches.
Acknowledgements
This work is supported by National Natural Science Foundation of China. NO.81100911
References
- Abrams MB, Nilsson I, Lewandowski SA, Kjell J, Codeluppi S, Olson L, Eriksson U. Imatinib enhances functional outcome after spinal cord injury. PLoS One 2012; 7: e38760.
- Fitch MT, Doller C, Combs CK, Landreth GE, Silver J.Cellular and molecular mechanisms of glial scarring and progressive cavitation: in vivo and in vitro analysis of inflammation-induced secondary injury after CNS trauma. J Neurosci 1999; 19: 8182-8198.
- Fitch MT, Silver J. CNS injury, glial scars, and inflammation: Inhibitory extracellular matrices and regeneration failure. Exp Neurol 2008; 209: 294-301.
- Gonias SL, Carmichael A, Mettenburg JM, Roadcap DW, Irvin WP, Webb DJ. Identical or overlapping sequences in the primary structure of human alpha(2)- macroglobulin are responsible for the binding of nerve growth factor- beta, platelet-derived growth factor-BB, and transforming growth factor- beta. J Biol Chem 2000; 275: 5826-5831.
- Heldin CH, Ostman A, Rönnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim Biophys Acta. 1998; 1378: F79-113.
- Heldin CH, Betsholtz C, Johnsson A, Nistér M, Ek B, Rönnstrand L, Wasteson A, Westermark B. Plateletderived growth factor: mechanism of action and relation to oncogenes. J Cell Sci Suppl. 1985; 3: 65-76.
- Caplan AI, Correa D. PDGF in bone formation and regeneration: new insights into a novel mechanism involving MSCs. J Orthop Res 2011; 29: 1795-1803.
- Hollinger JO, Hart CE, Hirsch SN, Lynch S. Friedlaender G.E., Recombinant human plateletderived growth factor: biology and clinical applications. J Bone Joint Surg Am. 1998; 90: 48-54.
- Sasahara M, Amano S, Sato H, Yang JG, Hayase Y, Kaneko M, Sato I, Suzaki M, Hazama F. Normal developing rat brain expresses a platelet-derived growth factor B chain (c-sis) mRNA truncated at the 5' end. Oncogene. 1998; 16: 1571-1578.
- Sasahara A, Kott JN, Sasahara M, Raines EW, Ross R, Westrum LE. Platelet-derived growth factor B-chainlike immunoreactivity in the developing and adult rat brain. Brain Res Dev Brain Res. 1992; 68: 41-53.
- Li XL, Zhou X, Wang XY, Zhang HT, Qin DX, Zhang H, Li Q, Li M, Wang TH. Temporal changes in the expression of some neurotrophins in spinal cord transected adult rats, Neuropeptides. 2007; 3: 135-143.
- Dow LE, Premsrirut PK, Zuber J, Fellmann C, McJunkin K, Miething C, Park Y, Dickins RA, Hannon GJ, Lowe SW. A pipeline for the generation of shRNA transgenic mice. Nat Protoc 2012; 7: 374-393.
- Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2000; 30; 1757-1766.
- Takimoto Y, Kuramoto A. Regulation of the human platelet-derived growth factor B-chain gene. Hiroshima J Med Sci. 1993; 42: 137-142.
- Valenzuela CF, Kazlauskas A, Weiner JL. Roles of platelet-derived growth factor in the developing and mature nervous systems, Brain Res Brain Res Rev. 1997; 24: 77-89.
- Nilsson J, Ksiazek T, Heldin CH, Thyberg J. Demonstration of stimulatory effects of platelet-derived growth factor on cultivated rat arterial smooth muscle cells. Differences between cells from young and adult animals. Exp Cell Res. 1983; 145: 231-237.
- Ross R. Platelet-derived growth factor. Annu Rev Med. 1987; 38: 71-79.
- Heldin CH. Platelet-derived growth factor—an introduction. Cytokine Growth Factor Rev 2004; 15:195-196.