Research Article - Journal of RNA and Genomics (2016) Volume 12, Issue 1
Comparative susceptibility of southern and western corn rootworm adults and larvae to vATPase-A and Snf7 dsRNAs
Adriano E Pereira1*, Newton P Carneiro2, and Blair D Siegfried31Department of Entomology, University of Nebraska, Lincoln, NE, USA
2Embrapa Maize and Sorghum, Road MG 424, Sete Lagoas, MG, Brazil
3Department of Entomology and Nematology, University of Florida, Gainesville, FL, USA
- Corresponding Author:
- Adriano E Pereira
Tel: (402) 499-3425
Fax: (402) 472-4687
E-mail: aelias374@yahoo.com.br
Received Date: 26 August 2016; Revised Date: 14 October 2016; Accepted Date: 24 October 2016; Published Date: 27 October 2016
Copyright: © First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Abstract
Corn rootworms (CRW) are the most important corn pests in the U.S. Corn Belt. Damage is caused by larval feeding on the plant roots. RNAi has been studied in CRW and has proven to be effective for the management of this insect with effects on both larval and adult stages. The objective of this study was to determine comparative susceptibility of adult and larva of Southern Corn Rootworm (SCR) and Western Corn Rootworm (WCR) to two different lethal RNAi target genes (vacuolar (v)-ATPase-A and Snf7). Adults and larvae were exposed to dsRNAs overlaid on artificial diet five times every other day. WCR larval susceptibility to vATPase-A was approximately 4-fold more tolerant than SCR, but were similar for Snf7 (2.6 ng/cm2 for both species). For adults, LC50s were generally higher for vATPase-A dsRNA relative to larvae with a 20-fold higher LC50 for WCR (SCR=33.3 ng/cm2; WCR=657.3 ng/cm2) relative to Snf7 dsRNAs (SCR=13.2 ng/cm2; WCR=60.2 ng/cm2). Gene silencing was also evaluated in adults and larvae exposed to the LC50 dsRNAs, and gene knockdown ranged from 48% up to 83% in adults fed for eight days in dsRNA, and from 55% to 89% in larvae fed for 24, 48 and 72 h, compared to control treatments. This is the first study to report LC50 values for dsRNA in WCR and SCR adults. The results indicate that both larvae and adults of both species were affected by vATPase-A and Snf7 dsRNAs suggesting that RNAi for the management of CRW should be considered for both stages.
Keywords
RNA interference, RNAi, Diabrotica undecimpunctata howardi, Diabrotica virgifera virgifera, Resistance monitoring, Corn rootworm
Introduction
Since its initial characterization in Caenorhabditis elegans (Maupas) nearly two decades ago, RNA interference (RNAi) has become a powerful genetic tool, allowing functional genomic studies in many organisms through gene knockdown after exposure to double-stranded (ds)RNA (Fire et al, 1998; Fire, 2007). RNAi was first documented through injection and soaking of C. elegans in dsRNA solution, causing knockdown of genes related to motility and to embryonic development (Fire et al, 1998; Timmons and Fire, 1998; Timmons et al, 2001). The use of RNAi has been shown in many studies to be effective in suppressing gene expression in insect pests from several orders (Whyard et al, 2009; Huvenne and Smagghe, 2010; Burand and Hunter, 2013). Identifying lethal target genes to control important pests such as CRW (Baum et al, 2007; Bolognesi et al, 2012; Chu et al, 2014; Khajuria et al, 2015; Fishilevich et al, 2016), cotton bollworm, Helicoverpa armigera (Hübner) (Mao et al, 2007, 2011), and Colorado potato beetle, Leptinotarsa decemlineata (Say) (Zhu et al, 2011; Palli, 2014; Whyard, 2015) has become an important focus of RNAi research. Some studies have involved genomic screenings to identify lethal target genes in important pests such as L. decemlineata (Swevers et al, 2013) and CRW (Baum et al, 2007). Transgenic plants that express short hairpin RNA molecules have been developed for controlling pests such as the CRW and additional studies have tested a variety of methods of exposure involving synthetic dsRNA molecules (Baum et al, 2007).
WCR, Diabrotica virgifera virgifera LeConte, is by far the most important diabroticite species to corn production throughout the U.S. Corn Belt (Gray et al, 2009; Meinke et al, 2009). WCR has been reported to evolve resistance to four major insecticide classes (Ball and Weekman, 1962; Meinke et al, 1998; Wang et al, 2013; Pereira et al, 2015) as well as behavioral resistance to crop rotation where gravid females leave corn fields to feed and oviposit in soybean fields (Levine et al, 2002; Curzi et al, 2012; Chu et al, 2013). More recently, resistance to Cry3Bb1 with cross-resistance to mCry3A and eCry3.1Ab has been reported (Gassmann et al, 2011; Wangila et al, 2015; Zukoff et al, 2016; Gassmann, 2016).
SCR, Diabrotica undecimpunctata howardi Barber is a polyphagous pest that occurs in the southern region of the US and that migrates to the Corn Belt during spring and summer. SCR can cause damage to a variety of plants especially corn, but also including peanuts, cucurbits, and soybeans in the southern region of the U.S. (Isely, 1929; Krysan, 1986). SCR and WCR cause significant damage and yield loss to corn plants as the larvae feed on corn roots affecting water and nutrient uptake, and heavy infestations can cause lodging due to loss of stability (Krysan, 1986; Urías-López and Meinke, 2001; Dun et al, 2010; Tinsley et al, 2013).
Current management strategies for CRW in the U.S. Corn Belt have relied extensively on transgenic plants expressing Bt toxins (for WCR) or insecticides applied in the soil for larval control or as adulticides to prevent egg laying of gravid females (Meinke et al, 2009). Given the challenges to rootworm control because of resistance evolution, it is critical to find new management approaches with different modes of action. RNAi has become a promising tool for insect control that may soon be available to growers in corn hybrids expressing dsRNA for corn rootworm control (Ahmad et al, 2016; Bachman et al, 2016). Baum et al, (2007) has shown efficient WCR larval control using plants expressing vATPase-A dsRNAs.
The most challenging factor for the success of RNAi technique is its delivery to the insect, which has been tested as feeding in artificial diet or in-planta in several insect pests with both success and failure (Huvenne and Smagghe, 2010; Yu et al, 2013). RNAi has been shown to be effective in some insect pests as adults, including WCR (Rangasamy and Siegfried, 2012) and L. decemlineata (Palli, 2014; Whyard, 2015). Because WCR adults feed on above ground plant structures such as leaves and especially silk and pollen, and because dsRNA can be expressed in these above ground tissues (Bachman et al, 2016), transgenic plants expressing RNAi traits could affect both larvae and adults (Rangasamy and Siegfried, 2012).
The genes used in this research target two housekeeping genes considered essential to cellular functions and have been shown to cause lethality in both rootworm species (Baum et al, 2007; Bolognesi et al, 2012; Bachman et al, 2013; Levine et al, 2015). Snf7 is a component of the endosomal sorting complex (ESCRTIII) in eukaryotic cells, is required for the sorting and transport of molecules and concentration of proteins in multivesicular bodies-MVB (Rusten et al, 2012). The Snf7 protein has multiple functions in midgut cells including autophagy, membrane stability, interaction with and recruitment of de-ubiquitinating enzymes to the ESCRT pathway, intraluminal vesicle formation in MVBs, and cytokinesis (Koci et al, 2014). The vacuolar (v)- ATPase subunit A gene encodes an important proton pump and pH regulator located in the cell membrane and participates in many biological processes in cells and organelles, including the regulation of pH of intracellular organelles, receptor-mediated endocytosis, intracellular targeting of lysosomal enzymes, membrane trafficking of molecules, protein degradation, homeostasis of cytoplasmic pH, and coupled transport of small molecules (Beyenbach and Wieczorek, 2006; Forgac, 2007).
The objective of this research was to assess and compare the susceptibility of adults and larvae of SCR and WCR to vATPase-A and Snf7 dsRNAs. We also assessed the gene knockdown after exposing those life stages to the calculated dsRNA LC50. Our results showed that WCR and SCR adults, along with SCR larvae, are more susceptible to Snf7 when compared to vATPase-A dsRNA, and that WCR adults possess reduced susceptibility to vATPase-A when compared to SCR adults. These results indicate that although adults of both WCR and SCR are less susceptible to RNAi than larvae, they may not be completely immune to the concentrations expressed by transgenic plants which could have implications to resistance management.
Material And Methods
Insects
SCR and non-diapause WCR adults and eggs were purchased from a commercial vendor (Crop Characteristics®, Farmington, MN). Adults were shipped overnight to the University of Nebraska-Lincoln, Insect Toxicology Laboratory, and maintained in BugDorm® cages (MegaView Science Co., Taichung, Taiwan) at 25°C and 60-80% RH and provided with artificial diet for 24 h before initiating bioassays. Eggs were received in petri dishes in fine soil and maintained in a chamber at 25°C and 70% RH until hatching.
dsRNA synthesis
A single adult beetle was homogenized in 1.5 mL microcentrifuge tube to extract the RNA with RNeasy kit (Qiagen, Valencia, CA) following manufacturer’s protocol. Then, 1μg of total RNA was used to synthesize cDNA using the QuantiTect reverse transcription kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. The PCR templates for in-vitro transcription of vATPase-A (accession # KX982002) and Snf7 (accession # KX982003) dsRNAs for SCR and vATPase-A (accession # CN498337.1) and Snf7 (Bolognesi et al, 2012) dsRNAs for WCR were amplified using gene-specific primers for each species with the T7 polymerase promoter sequence, which amplified 289 bp and 258 fragments for vATPase-A and 319 bp and 289 bp fragments for Snf7, respectively (Table 1). After confirming template sequences for SCR and WCR, specific vATPase-A and Snf7 were synthesized in-vitro using MEGAscript high-yield transcription kit (Applied Biosystems Inc., Foster City, CA), purified using RNAeasy Mini kit (Qiagen) following the manufacturer’s protocol and stored at -20°C (Rangasamy and Siegfried, 2012). Green Fluorescent Protein (GFP) dsRNA that amplified 370 bp (Table 1) and water were used as negative controls.
| Species/ gene | Primer sequences for dsRNA synthesis | PL (bp) |
|---|---|---|
| WCR | ||
| vATPase-A | F: TAATACGACTCACTATAGGGTATTGTACAGGTG R: TAATACGACTCACTATAGGGCAATTTCCAAG |
258 |
| Snf7 | F: TAATACGACTCACTATAGGGTTGCACTCCAAGCCCTCAAAA R: TAATACGACTCACTATAGGGTTCCAGATCGTCGGTGAATCC |
289 |
| GFP | F: TAATACGACTCACTATAGGGGGTGATGCTACATACGGAAAG R: TAATACGACTCACTATAGGGTTGTTTGTCTGCCGTGAT |
370 |
| SCR | ||
| vATPase-A | F: TAATACGACTCACTATAGGGTATTGTACAGGTG R: TAATACGACTCACTATAGGGCAATTTCCAAG |
289 |
| Snf7 | F: TAATACGACTCACTATAGGGATGTTGCACTCCAAGCCCTCAAAA R: TAATACGACTCACTATAGGGGTCCAGATCGTCGGTGAATCC |
319 |
| GFP | F: TAATACGACTCACTATAGGGGGTGATGCTACATACGGAAAG R: TAATACGACTCACTATAGGGTTGTTTGTCTGCCGTGAT |
370 |
Table 1: Primer sequences for WCR and SCR Snf7, vatpase-A, and GFPdsrnas.
Quantitative real-time PCR (qPCR)
For measurement of gene knockdown, similar exposure conditions were employed. For larval assays, SCR and WCR neonates were exposed to the calculated LC50 of dsRNA for 24, 48 or 72 h for both genes in artificial diet pellets as described for susceptibility bioassays, and flash frozen at -80°C. qPCR assays were performed with three biological replicates including three samples of 20-30 larvae per treatment in each biological replicate. Three determinations of gene expression were conducted for each sample. For gene knockdown of adults, beetles were exposed four times every other day to the calculated LC50 dsRNA of each gene on artificial diet as described for susceptibility bioassays and flash frozen at -80°C after eight days of continuous feeding on dsRNA. qPCR assays were performed with two biological replicates, three samples per biological replicate, and three technical replicates per sample with each sample containing two beetles. Larval exposure to dsRNA was performed for 72 h to minimize microbial contamination.
Neonates and adults were collected and flash-frozen in liquid nitrogen. RNA was extracted with RNeasy kit (Qiagen, Valencia, CA) following the manufacturer’s protocol, and stored at -80°C. Total RNA (500 ng) was used to synthesize first strand cDNA using the Quantitech Reverse Transcription kit (Qiagen, Valencia, CA). The concentration of extracted RNA was estimated using a NanoDrop1000 (Thermo-Fisher Scientific, Wilmington, DE, USA), and quality evaluated on 1% agarose gel electrophoresis. qPCR primers were generated in primer3 website (http://primer3.ut.ee/) (Table 2). β-actin was used as reference gene to compare with Snf7 and vATPase-A (Table 2). For controls in both larvae and adult bioassays, we used GFP dsRNA at the respective LC50 concentration for each gene, as well as double distilled water.
| Species/ gene | Primer sequences for qPCR | PL (bp) | Slope | R2 | Eff. (%) |
|---|---|---|---|---|---|
| WCR | |||||
| β-Actin | F: TCCAGGCTGTACTCTCCTTG R: CAAGTCCAAACGAAGGATTG |
134 | -3.419 | 0.999 | 96.1 |
| vATPase-A | F: GGAAGAAGATGATCTAGCCGAAATT R: TTGTCCGTTTCTGCCAGAGA |
67 | -3.361 | 0.993 | 98.4 |
| Snf7 | F: TTACGAGGCCCAGGCTTCC R: CCGACGATCTGGATGACGA |
206 | -3.533 | 0.974 | 91.9 |
| SCR | |||||
| β-Actin | F: CCAGCTGCTTCCATACCCAA R: TGCCAGTTCCAGTTCCCTAG |
129 | -3.452 | 0.996 | 94.8 |
| vATPase-A | F: GGAAGAAGATGATCTAGCCGAAATT R: TTGTTCGTTTCTGCCAGAGA |
67 | -3.181 | 0.997 | 106.2 |
| Snf7 | F: CCGACGATCTGGACGATGA R: TTATGAGGCCCAGGCTTCC |
205 | -3.151 | 0.982 | 107.7 |
Table 2: Primer sequences for WCR and SCR Snf7,vatpase-A, and β-actinqpcr.
qPCR analysis was performed using SYBR green kit (Applied Biosystems Inc., Foster City, CA) and the 7500 Fast System real-time PCR detection system (Applied Biosystems Inc., Foster City, CA). qPCR cycling parameters included 40 cycles of 95°C for 3 seconds, 58°C for 30 seconds, as described in the supplier’s protocol (Applied Biosystems Inc., Foster City, CA). At the end of each PCR reaction, a melting curve was generated to confirm single peak and rule out the possibility of primer-dimer and non-specific product formation. Relative quantification of the transcripts were calculated using the comparative 2-ddCt method (Livak and Schmittgen, 2001) and were normalized to β-actin (Rangasamy and Siegfried, 2012).
Bioassays
For larval and adult bioassays, 48-well plates (Costar # 3548, Corning Incorporated, Corning, NY) were used for exposure to artificial diet and sealed with plate-sealer (Thermo Fisher Scientific, Rochester, NY) with one hole punched per well for larvae and four holes punched per well for adults using # 1 insect pin. The artificial diet used for the adults of both species was slightly modified from Branson and Jackson (1988) and described in Khajuria et al, (2015). SCR larval diet was purchased from Frontier Agricultural Sciences (Newark, DE) and for WCR larval bioassay, we used a proprietary artificial diet provided by Dow AgroSciences (Tan et al, 2016). The dsRNA concentrations used to estimate LC50 values for adults and larvae included 10X series diluted from 1000 to 0.1 ng/cm2,and for WCR adult susceptibility bioassay with vATPase-A, we included the concentration 5000 ng/cm2 to achieve 100% mortality, as 1000 ng/cm2 caused only 60% mortality. For SCR and WCR adult bioassays, one adult was placed per well containing 2 adult artificial diet pellets (4 mm diameter, 2 mm height; area=0.1256 cm2) each treated with 3 μL of the respective dsRNA concentration, and each concentration was replicated four times for a total of 32 adults/treatment.
For SCR larval bioassays, two neonates were transferred per well containing two larval artificial diet pellets (same dimensions as for adults) each treated with 3 μL of the respective dsRNA concentrations and each concentration was replicated three times, for a total of 48 larvae/treatment. For WCR larval bioassays, only one neonate was placed per well for a total of 24 larvae/treatment. For WCR larval assays, mortality on the diet pellets was consistently >20% and the larvae did not readily accept the diet pellets; therefore, plates contained 200 μL of WCR larval artificial diet treated with 40 μL of respective dsRNA concentration, so that the larvae could be in contact with diet and feed all the time. Each bioassay was performed twice using different cohorts of larvae and adults on different days. Adult mortality was recorded after 14 days and larval mortality, after 12 days. Only those data in which control mortality was < 20% were included in analyses.
To minimize microbial contamination of diet and to make the bioassays standardized, neonates and adults from each treatment were carefully transferred to new plates containing new treated diet every other day, with five exposures to dsRNA over 10 days. After the 5th dsRNA exposure, untreated artificial diet was provided. For WCR larval bioassays, larvae were transferred after six days to freshly treated diet. Artificial diet pellets in the control treatments were treated with 3 μL of double distilled water, and WCR larval diet was treated with 40 μL of double distilled water. To facilitate transfer to new diet, adults were anesthetized at 4°C for 15 min to allow easy transfer to the new plates and minimize stress.
Statistical analysis
PoloPlus-PC software (LeOra, 1987) probit analysis was used to analyze the mortality and generate the LC50s (Finney, 1971). PoloPlus corrects for control mortality automatically using Abbott’s formula (Abbott, 1925). Relative gene expression for larvae and adults were analyzed with an ANOVA and compared by least square means and Tukey adjustments at p <0.05, using PROC GLIMMIX in SAS.
Results
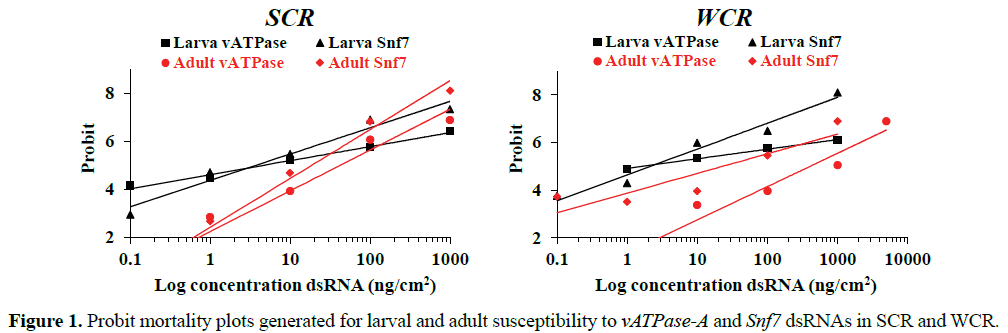
Results of lethal dsRNA bioassays for both species and gene targets are presented in Table 3. Mortality plots of logarithmic concentrations were generated in Probit analysis to illustrate the differences in susceptibility between larvae and adults to dsRNA in SCR and WCR (Figure 1). The plots indicate the general fit of the data and that the larvae exhibited much shallower slopes relative to the adults. These plots may reflect differences in feeding behavior of the two stages that ultimately affects overall exposure and response.
SCR vATPase-A LC50s for adults and larvae were 33.3 and 8.0 ng/cm2, respectively, and 13.2 and 2.6 ng/cm2 for Snf7, respectively (Table 3). WCR vATPase-A LC50s for adults and larvae were 657.3 and 1.70 ng/cm2 respectively, and for Snf7, 60.2 and 2.62 ng/cm2, respectively (Table 3). In general, WCR adults exhibited higher LC50s for both genes when compared to SCR, with 20-fold higher LC50 for v-ATPase-A dsRNA and 4.5- fold higher for Snf7 dsRNA.
| Species | Life stage | Target gene |
N* | Slope (± SE) |
LC50 (± CI) (ng/cm2) |
X2 (d.f.) |
|---|---|---|---|---|---|---|
| SCR | Adult | Snf7 | 384 | 1.35 (0.55) | 13.2 (5.0-20.0) | 5.4 (3) |
| SCR | Larval | Snf7 | 576 | 1.04 (0.09) | 2.6 (0.9-6.0) | 5.0 (3) |
| SCR | Adult | vATPase-A | 384 | 1.60 (0.21) | 33.3 (5.4-100.5) | 8.5 (3) |
| SCR | Larval | vATPase-A | 576 | 0.65 (0.07) | 8.0 (3.9-14.7) | 1.3 (3) |
| WCR | Adult | Snf7 | 384 | 1.54 (0.23) | 60.2 (21.9-120.4) | 3.1 (3) |
| WCR | Larval | Snf7 | 288 | 1.41 (0.17) | 2.62 (0.23-8.75) | 5.0 (3) |
| WCR | Adult | vATPase-A | 448 | 1.59 (0.25) | 657.3 (148.5-1406.3) | 8.7 (4) |
| WCR | Larval | vATPase-A | 288 | 0.41 (0.14) | 1.70 (0.01-10.6) | 0.04 (2) |
| WCR | Larval | Snf7 | - | - | 1.20** | - |
| WCR | Larval | vATPase-A | - | - | 1.82** | - |
Table 3: Comparative susceptibility (ng/cm2) of larvae and adults of WCR and SCR laboratorycolonies, exposed to Snf7 and vatpase-Adsrnas five times every other day.
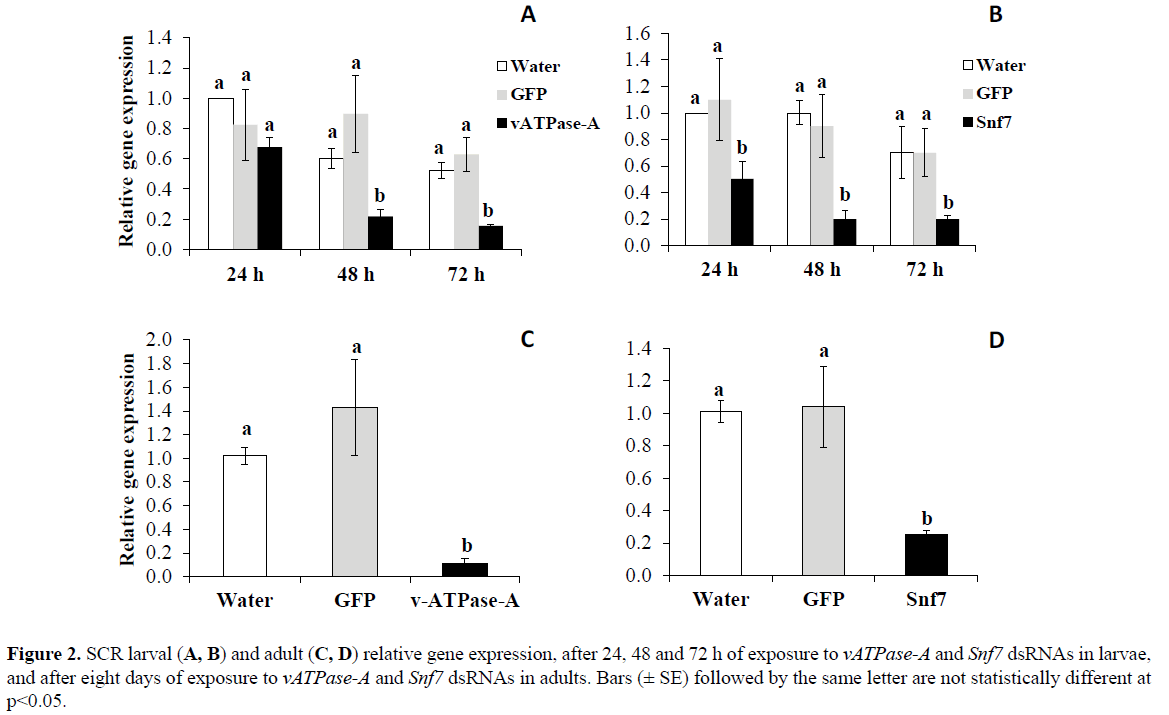
Results from qPCR analysis indicate that SCR neonates fed on artificial diet treated with either Snf7 or vATPase-A dsRNA LC50s for 24, 48, and 72 h showed substantial reductions in transcript levels for both genes. Significantly reduced expression compared to control water was observed at 48h (62%) and 72 h (70%) for vATPase-A (p< 0.0001), and 55, 80, and 71% for Snf7 (p< 0.0001), respectively (Figure 2). Significant differences were detected in adults fed with artificial treated diet with either vATPase-A (p< 0.001) or Snf7 (p< 0.01) dsRNA LC50s for eight days, with reduction in transcript levels of 92% and 76% for vATPase-A and Snf7, respectively, relative to the control treatments (Figure 2).
Figure 2: SCR larval (A, B) and adult (C, D) relative gene expression, after 24, 48 and 72 h of exposure to vATPase-A and Snf7 dsRNAs in larvae, and after eight days of exposure to vATPase-A and Snf7 dsRNAs in adults. Bars (± SE) followed by the same letter are not statistically different at p<0.05.
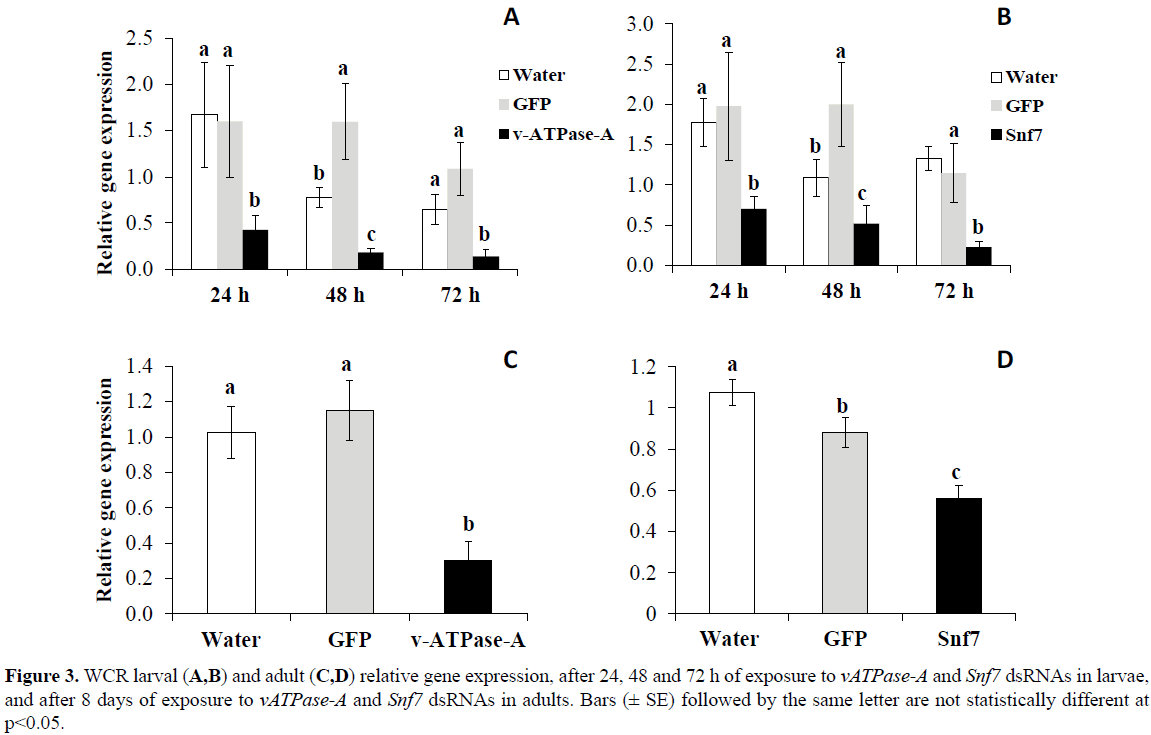
For WCR, similar results were found after neonates fed on artificial diet treated with either vATPase-A or Snf7 dsRNA LC50s. Significant differences were observed in the reduction of transcript levels relative to the controls at 24, 48 and 72 h, of 73, 89, and 87% for vATPase-A (p< 0.001), respectively, and of 61, 74, and 83% for Snf7 (p< 0.05), respectively (Figure 3). Significant differences were also detected in WCR adults fed on artificial diet treated with either vATPase-A (p< 0.001) or Snf7 (p< 0.0001) dsRNA LC50s for eight days with reduction in transcript levels of 74% for vATPase-A and 48% for Snf7 relative to the control treatments (Figure 3).
Discussion
To date, this is the first study to present LC50s of lethal dsRNAs for adults and larvae of two closely related species. The susceptibility of SCR and WCR larvae to Snf7 dsRNA was similar for both species (calculated LC50 for both species, 2.6 ng/cm2). In contrast, adult WCR were approximately 5-fold more tolerant to Snf7 dsRNA than SCR at the Bolognesi R, Ramaseshadri P, Anderson . For vATPase-A dsRNA in larvae, the SCR LC50 was >4-fold higher than WCR LC50, although 95% confidence intervals overlapped. In contrast to results with Snf7, susceptibility of WCR adults to vATPase-A dsRNA changed dramatically between larvae and adults, with the LC50 of adults nearly 400-fold higher than in larvae (Table 3).
These results indicate that there are differences in susceptibility between closely related species and between life stages for dsRNAs, targeting the same gene. These differences should be taken into consideration when exploring RNAi approaches for the control of CRW complex. Although the target life stage for lethal RNAi technologies will be larvae, which are also generally more susceptible than adults, our results suggest that these technologies my not be harmless to adults. In contrast to Bt toxins which do not affect adults either because the gut environment differs from larvae or because the molecular target is not expressed in adults (Nowatzki et al, 2006), it is clear that lethal dsRNA has the potential to adversely affect rootworm adults. This could be especially important if selection is directed against both larval and adult stages. Evaluating the expression of dsRNA in aboveground tissues of the corn plants may be an important factor in addressing resistance potential for this new technology.
The differences in susceptibility between larval and adult SCR were not as apparent as in WCR, with LC50s around 4-fold higher in adults when compared to larvae (Table 3) although the slopes were again consistently lower for larvae. It should be noted that differences in WCR susceptibility between adults and larvae could be due to differences in artificial diet for each life stage, given that larval diet becomes contaminated by microbial growth faster than adult diet. Adult bioassays showed minimal contamination problem as beetles usually consume most of the diet pellet before contamination.
It has been previously reported that WCR adults are affected by vATPase-A dsRNA at concentrations of 500 ng/diet pellet (3,981 ng/cm2) and 1,000 ng/pellet (7,961 ng/cm2) with mortality reaching 95% after 15 days of exposure (Rangasamy and Siegfried 2012). These concentrations are comparable to our results (LC90=4,230 ng/cm2), with LC50 of 657.3 ng/cm2 (Table 3). WCR adults were also shown to be less susceptible to Snf7 dsRNA (60.2 ng/cm2) with 4.5-fold higher LC50 when compared to SCR adults (13.3 ng/cm2).
Koci et al, (2014) reported that silencing Snf7 gene in WCR neonate midgut’s cells is extremely lethal, causing disintegration and macroautophagy of the cell membranes. Although the symptomatology of vATPase-A dsRNA acting in midgut cells of any insect has not been reported, one can assume that there should be differences in the effects between the two dsRNAs in the cells, based on the role of each gene as described in introduction. In the bioassays, it was observed that mortality of both WCR and SCR adults to Snf7 dsRNA started around 4-6 days after initial exposure. In contrast, mortality by vATPase-A dsRNA was first observed one to two days later, at 5-7 days after initial exposure. Interestingly, adults of both species exposed to higher concentrations of Snf7 dsRNA caused the sudden onset of death after 4-6 days. In contrast, adults exposed to vATPase-A dsRNA exhibited sluggishness and lack of coordinated mobility prior to mortality. Therefore, it is assumed that Snf7 appears to be more potent in terms of symptomatology, which again may be translated by the lower LC50s for both species, when compared to vATPase-A.
It is of interest to note that there was not a strong correlation between the level of knockdown and toxicity. In general, vATPase-A exhibited significantly greater knockdown in both adults and larvae, but was consistently less toxic than Snf7. A number of factors could contribute to this discrepancy such as tissue specific expression.
WCR larval susceptibility to vATPase-A (1.70 ng/cm2) and Snf7 (2.62 ng/cm2) dsRNAs in this study is comparable to the LC50s (1.20 and 1.82 ng/cm2, respectively) reported by Baum et al, (2007), although confidence intervals were not previously reported. Bolognesi et al, (2012) also calculated baseline susceptibility of both WCR and SCR neonates exposed to WCR Snf7 dsRNA (240 bp fragment) incorporated into artificial diet and observed a 3.5-fold difference in LC50s for both species (4.3 and 1.2 ng dsRNA/mL of diet in WCR and SCR, respectively). Levine et al, (2015) reported the LC50 of Snf7 dsRNA specific to WCR to range between 3.5 and 9.2 ng/mL of diet for SCR. Preliminary studies were performed to determine the specificity of Snf7 and vATPase-A dsRNAs in adults between WCR and SCR, and also observed similar sensitivity of WCR and SCR to heterospecific dsRNAs for both genes, as they show >97% gene sequence identity (data not shown).
Significant knockdown of expression was evident for both Snf7 and vATPase-A transcripts in both larvae and adults of both species, with the exception of SCR neonates exposed to the vATPase-A dsRNA LC50 (2.6 ng/cm2) for 24 h. The results with larvae are consistent with other studies that demonstrate gene silencing in WCR neonates 24 h after dsRNA ingestion (Bolognesi et al, 2012). There were minimal differences in percentage of reduction in transcript levels among the different feeding times on dsRNA, again except for SCR neonates feeding in vATPase-A dsRNA LC50 for 24 h. Gene knockdown was clearly observed for both genes in both species, suggesting that CRW are readily affected by dsRNA in both developmental stages.
It should be noted that the level of gene expression after ingestion of dsRNA did not necessarily correlate with the level of mortality. The percentage of gene knockdown was consistently higher for vATPase-A than for Snf7 although in general, Snf7 caused consistently higher mortality. There were also inconsistencies between larvae and adults especially with vATPase-A and WCR where adults were much less sensitive than larvae although knockdown was comparable. These results further support the contention that each targeted gene behaves differently and that many factors contribute to a lethal response other than simply the percentage of gene knockdown. For example, Snf7 expression may be significantly reduced in midgut tissue causing high levels of mortality (Koci et al, 2014), but because the expression was determined from whole bodies, lower knockdown was observed. In addition, the relative roles of the two genes in larvae and adults may change because of different requirements for growth and development. Assessing the relationship between exposure, mortality and gene knockdown could provide a more accurate assessment of how these factors interact.
Although it is not possible to accurately compare susceptibility on a dose/body weight basis because the amount of dsRNA consumed cannot be accurately quantified, adults are more than 200-fold heavier than neonate larvae and are likely to consume less diet on a per body weight basis. Therefore, if relative susceptibility could be determined on a dose/body weight basis, the conclusion that larvae are more susceptible than adults might be inaccurate. Studies designed to estimate the amount of dsRNA ingested by each life stage will provide a basis for future research to understand the dsRNA threshold that affects SCR and WCR life stages by RNAi.
Conclusions
This study is an important first-step in assessing WCR and SCR susceptibility to RNAi in adults and larvae and show that adults may potentially be affected by exposure to transgenic maize plants that express RNAi traits. In addition, the methods for determining adult and larval susceptibility to lethal dsRNA provide a foundation for monitoring efforts designed to detect changes in susceptibility that may be the result from selection pressure by the use of RNAi plants in near future. In addition, identifying the expression of dsRNA in above ground tissue of the corn plant is necessary to predict the effects of dsRNA on adults and to make appropriate recommendations to delay and/or mitigate WCR resistance evolution to this new mode of action.
Improvements in artificial diets for neonate bioassays need to be addressed to allow faster and less laborious bioassays. If selection for resistance occurs for both developmental stages, such selection should be considered in WCR resistance management strategies. Feeding bioassays involving exposure to transgenic plants expressing RNAi traits with adults should be performed for possible effects on adults. Also, the results are important for possible studies of sublethal exposures, in order to elucidate the potential impact on fitness parameters after exposure to RNAi in SCR and WCR and subsequent effects at the population level.
Acknowledgements
The authors are thankful to Dr. Sek Yee Tan from Dow AgroSciences, for providing WCR larval diet for the bioassays, and to the valuable comments from anonymous reviewers. The project has been funded by Department of Entomology/ University of Nebraska-Lincoln/Institute of Agriculture and Natural Resources, as an assistantship awarded to the first author.
Competing Interests
The authors have no competing interests.
References
- Abbott W. 1925. A method of computing the effectiveness of an insecticide. J Econ Entomol, 18, 265-267.
- Ahmad A, Negri I, Oliveira W et al. 2016. Transportable data from non-target arthropod field studies for the environmental risk assessment of genetically modified maize expressing an insecticidal double-stranded RNA. Trans Res, 25, 1-17.
- Bachman PM, Bolognesi R, Moar WJ et al. 2013. Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (DiabroticavirgiferavirgiferaLeConte). Trans Res, 22, 1207-1222.
- Bachman PM, Huizinga KM, Jensen PD et al. 2016. Ecological risk assessment for DvSnf7 RNA: A plant-incorporated protectant with targeted activity against western corn rootworm. RegulToxicolPharmacol, 81, 77-88.
- Ball HJ, and Weekman GT. 1962. Insecticide resistance in the adult western corn rootworm in Nebraska. J Econ Entomol, 55, 439-441.
- Baum JA, Bogaert T, Clinton W et al. 2007. Control of coleopteran insect pests through RNA interference. Nat Biotech, 25, 1322-1326.
- Beyenbach KW and Wieczorek H. 2006. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J ExpBiol, 209, 577-589.
- Bolognesi R, Ramaseshadri P, Anderson J et al. 2012. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (DiabroticavirgiferavirgiferaLeConte).PLoS One, 7, e47534.
- Branson T and Jackson J. 1988. An improved diet for adult Diabroticavirgiferavirgifera (Coleoptera: Chrysomelidae). J KansEntomolSoc, 61, 353-355.
- BurandJP, and Hunter WB. 2013. RNAi: Future in insect management. J InvertebrPathol, 112, S68-S74.
- Chu CC, Spencer JL, Curzi MJ, Zavala JA and Seufferheld MJ. 2013. Gut bacteria facilitate adaptation to crop rotation in the western corn rootworm. Proc Nat AcadSci, 110, 11917-11922.
- Chu CC, Sun W, Spencer JL, Pittendrigh BR, and Seufferheld MJ. 2014. Differential effects of RNAi treatments on field populations of the western corn rootworm. Pest BiochemPhysiol, 110, 1-6.
- Curzi MJ, Zavala JA, Spencer JL and Seufferheld MJ. 2012. Abnormally high digestive enzyme activity and gene expression explain the contemporary evolution of a Diabrotica biotype able to feed on soybeans. EcolEvol, 2, 2005-2017.
- Dun Z, Mitchell P andAgosti M. 2010. EstimatingDiabroticavirgiferavirgifera damage functions with field trial data: applying an unbalanced nested error component model. J ApplEntomol, 134, 409-419.
- Finney DJ. 1971. Probit analysis: Cambridge University Press, Cambridge, UK, 3rd edition.
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE and Mello CC. 1998.Potent and specific genetic interference by double-stranded RNA in Caenorhabditiselegans. Nat, 391, 806-811.
- Fire AZ. 2007. Gene silencing by double‐stranded RNA (Nobel Lecture).AngewChemInt Ed, 46, 6966-6984.
- Fishilevich E, VélezAM, Storer NP et al. 2016. RNAi as a management tool for the western corn rootworm, Diabroticavirgiferavirgifera. Pest ManagSci, In press.
- Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol, 8, 917-929.
- Gassmann AJ. 2016. Resistance to Bt maize by western corn rootworm: insights from the laboratory and the field. CurrOpin Insect Sci, 15, 111-115.
- Gassmann AJ, Petzold-Maxwell JL, Keweshan RS and Dunbar MW. 2011. Field-evolved resistance to Bt maize by western corn rootworm. PLoS One., 6, e22629.
- Gray ME, Sappington TW, Miller NJ, Moeser J and Bohn MO. 2009. Adaptation and invasiveness of western corn rootworm: intensifying research on a worsening pest. Annu Rev Entomol, 54, 303-321.
- Huvenne H and Smagghe G. 2010. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol, 56, 227-235.
- Isely D. 1929.The southern corn rootworm. Ark AgrExpSta, 232, 31p.
- Khajuria C, Velez AM, Rangasamy M et al. 2015. Parental RNA interference of genes involved in embryonic development of the western corn rootworm, DiabroticavirgiferavirgiferaLeConte. Insect BiochemMolecBiol, 63, 54-62.
- Koci J, Ramaseshadri P, Bolognesi R, Segers G, Flannagan R and Park Y. 2014. Ultrastructural changes caused by Snf7 RNAi in larval enterocytes of western corn rootworm (Diabroticavirgiferavirgifera Le Conte). PLoS One, 9, e83985.
- Krysan JL. 1986. Introduction: biology, distribution, and identification of pest Diabrotica. In Krysan JLM and Thomas A (Eds) Methods for the study of pest Diabrotica, Springer, New York, First Edition, pp 1-23.
- LeOra. 1987. POLO-PC: a user's guide to probit or logitanalysis.:LeOra Software Berkeley, CA.
- Levine E, Spencer JL, Isard SA, Onstad DW and Gray ME. 2002. Adaptation of the western corn rootworm to crop rotation: evolution of a new strain in response to a management practice. Am Entomol, 48, 94-107.
- Levine SL, Tan J, Mueller GM, Bachman PM, Jensen PD and Uffman JP. 2015. Independent action between DvSnf7 RNA and Cry3Bb1 protein in southern corn rootworm, Diabroticaundecimpunctatahowardi and Colorado potato beetle, Leptinotarsadecemlineata. PLoS One, 10, e0118622.
- Livak KJ and Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) method. Methods, 25, 402-408.
- Mao,YB, Cai WJ, Wang JW et al. 2007. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol, 25, 1307-1313.
- Mao YB, Tao XY, Xue XY, Wang LJ and Chen XY. 2011. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Trans Res, 20, 665-673.
- Meinke LJ, Sappington TW, Onstad DW et al. 2009. Western corn rootworm (DiabroticavirgiferavirgiferaLeConte) population dynamics.Agric Forest Entomol, 11, 29-46.
- Meinke LJ, Siegfried BD, Wright RJ and Chandler LD. 1998. Adult susceptibility of Nebraska western corn rootworm (Coleoptera: Chrysomelidae) populations to selected insecticides. J Econ Entomol, 91, 594-600.
- Palli SR. 2014. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. CurrOpin Insect Sci, 6, 1-8.
- Pereira AE, Wang H, Zukoff SN, Meinke LJ, French BW and Siegfried BD. 2015.Evidence of field-evolved resistance to bifenthrin in western corn rootworm (DiabroticavirgiferavirgiferaLeConte) populations in western Nebraska and Kansas.PLoS One, 10, e0142299.
- Rangasamy M and Siegfried BD. 2012. Validation of RNA interference in western corn rootworm DiabroticavirgiferavirgiferaLeConte (Coleoptera: Chrysomelidae) adults. Pest ManagSci, 68, 587-591.
- Rusten TE, Vaccari T and Stenmark H. 2012. Shaping development with ESCRTs. Nat Cell Biol, 14, 38-45.
- Swevers L, Huvenne H, Menschaert G et al. 2013. Colorado potato beetle (Coleoptera) guttranscriptome analysis: expression of RNA interference‐related genes. Insect MolBiol, 22, 668-684.
- Tan SY, Rangasamy M, Wang H et al. 2016. RNAi induced knockdown of a cadherin-like protein (EF531715) does not affect toxicity of Cry34/35Ab1 or Cry3Aa to Diabroticavirgiferavirgifera larvae (Coleoptera: Chrysomelidae). Insect BiochemMolBiol, 75, 117-124.
- Timmons L, Court DL and Fire A. 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditiselegans. Gene, 263, 103-112.
- Timmons L and Fire A. 1998. Specific interference by ingested dsRNA.Nat, 395, 854-854.
- Tinsley N, Estes R and Gray, M. 2013.Validation of a nested error component model to estimate damage caused by corn rootworm larvae. J ApplEntomol, 137, 161-169.
- Urías-López MA and Meinke LJ. 2001. Influence of western corn rootworm (Coleoptera: Chrysomelidae) larval injury on yield of different types of maize. J Econ Entomol, 94, 106-111.
- Wang H, Coates BS, Chen H, Sappington TW, Guillemaud T and Siegfried BD. 2013. Role of a gamma-aminobutryic acid (GABA) receptor mutation in the evolution and spread of Diabroticavirgiferavirgifera resistance to cyclodiene insecticides. Insect MolBiol, 22, 473-484.
- Wangila DS, Gassmann AJ, Petzhold-Maxwell JL, French BW and Meinke LJ. 2015. Susceptibility of Nebraska western corn rootworm (Coleoptera: Chrysomelidae) populations to Bt corn events. J Econ Entomol, 108, 742-751.
- Whyard S. 2015. Insecticidal RNA, the long and short of it. Science, 347, 950-951.
- Whyard S, Singh AD and Wong, S. 2009. Ingested double-stranded RNAs can act as species-specific insecticides. Insect BiochemMolBiol, 39, 824-832.
- Yu N, Christiaens O, Liu J et al. 2013. Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci, 20, 4-14.
- Zhu F, Xu J, Palli R, Ferguson J and Palli, SR. 2011.Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsadecemlineata. P ManagSci, 67, 175-182.
- Zukoff SN, Ostlie KR, Potter B et al. 2016. Multiple assays indicate varying levels of cross resistance in Cry3Bb1-selected field populations of the western corn rootworm to mCry3A, eCry3. 1Ab, and Cry34/35Ab1. J Econ Entomol, in press.