- Biomedical Research (2014) Volume 25, Issue 3
Characterization of SNPs in Oxidised Low Density Lipoprotein Receptor 1 (OLR1) gene in Mehsana and Banni breeds of buffalo.
C.V. Jawale1, Nadeem Shabir*1, C.D. Bhong1, A.K. Tripathi1, Patel Hiren1, D.N. Rank1,2, C.G. Joshi1,21Department of Animal Biotechnology,College of Veterinary Science and Animal Husbandry, Anand Agricultural University, Anand, Gujarat, India
2Department of Animal Genetics and Breeding,College of Veterinary Science and Animal Husbandry, Anand Agricultural University, Anand, Gujarat, India
- *Corresponding Author:
- Nadeem Shabir
Department of Animal Biotechnology
College of Veterinary Sciences & Animal Husbandry
Anand Agricultural University
Anand Gujarat,India
Accepted January 26 2014
Abstract
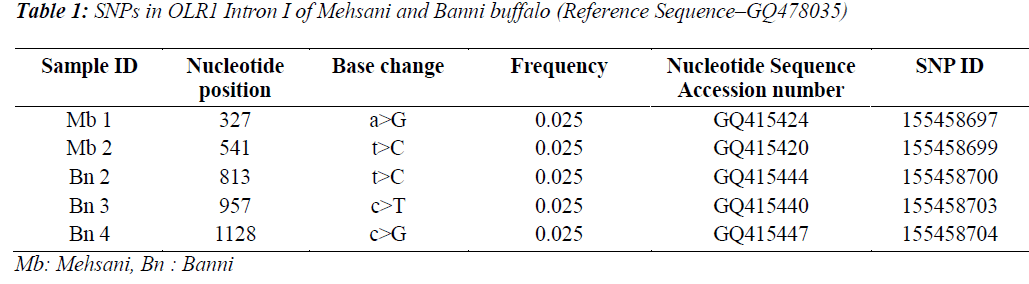
Single Nucleotide polymorphisms (SNP) are the most common type of DNA sequence polymorphisms and of mutation in vertebrates. Keeping in view the involvement of Oxidised Low Density Lipioprotein Receptor 1 (OLR1) gene in lipid metabolism and the higher milk fat percentages in Bubalus bubalis, the present study was carried out to explore the SNPs in OLR 1 intron I in Mehsani and Banni breeds of buffalo by gene cloning and sequencing. Five SNPs at nucleotide positions viz. 327, 541, 957 and 1128 were detected and all the SNPs followed breed lineage. The SNP at positions 327 and 541 were found in Mehsana breed alone while as rest of the explored SNPs restricted to Banni breed of Bubalus bubalis. The frequency of each SNP was found to be 0.025.
Keywords
Single Nucleotide Polymorphisms, buffalo, Oxidised Low density Lipoprotein Receptor 1.
Introduction
Oxidised Low Density lipoprotein Receptor 1 (OLR1) is the major protein that binds, internalizes, and degrades oxidized low-density lipoprotein. The role of OLR1 in lipid metabolism and the results of previous whole genome scan studies prompted the investigation of OLR1 as a candidate gene affecting milk composition traits [1]. In another study conducted in Dutch Holstein-Friesian cattle population in OLR 1 gene to estimate genotype effects on milk production traits revealed that OLRg.8232C>A had a significant effect (P < 0.05) on milk-fat percentage [2].
The oxidized form of the low-density lipoprotein (oxLDL) is involved in endothelial cell injury, dysfunction, and activation, all of which are implicated in the development of atherosclerosis [3]. It has been shown that oxLDL and its lipid constituents have numerous damaging effects on secretory activities of the endothelium, including induction of apoptosis [4]. The major protein that binds, internalizes, and degrades oxLDL, oxidized LDL receptor 1 (OLR1), was initially identified in bovine aortic endothelial cells [5]. In addition to binding oxLDL, OLR1 removes aged and apoptotic cells from blood circulation (Oka et al., 1998). The bovine OLR1 gene encodes 270 AA and has 72% identity to the human protein [6]. It has been previously demonstrated that the human OLR1 contains 6 exons, of which the first 3 corresponded to the Nterminal cytoplasmic, transmembrane, and connecting neck domains, and the last 3 encoded the lectin domain [7]. The genomic sequence of bovine OLR1, recently released by Baylor College of Medicine, contains 5 exons {GenBank accession no. NW_215807}.
Based on the aforementioned studies indicating the role of OLR1 in lipid metabolism and its correlation with milk fat percentage, the study of SNP exploration in this gene in Bubalus bubalis was initiated considering the higher milk fat percentages in buffaloes.
Materials and Methods
Sampling and DNA extraction
Blood samples, ten each from two breeds of buffalo, Mehsani (from Dudhsagar Dairy, Mehsana, Gujarat) and Banni (from field animals of Kachchh district, Gujarat) were collected at random from non related animals. Genomic DNA was extracted by Phenol:Chloroform method. The concentration and the purity of the DNA obtained were assessed by spectrophotometery and electrophoresis in 0.8% agarose gel respectively.
PCR amplification
The genomic DNA from each individual was amplified by PCR. A pair of primers designed using the reference sequence (Accession no. NC_007303.3) of OLR1 of Bos taurus (Forward:AGAAAACTACAATGCCTGGGT & Reverse: CACCACAAGGCAGAGAGT) was used to amplify a gene segment of 1163bp. Approximately 30ng of genomic DNA was amplified in 25μl PCR reaction consisting of 2X Master mix (MBI Fermentas). Thermal cycling was performed on Veriti ( Applied Biosystems) with an initial denaturing step at 950C for 10 mins followed by 40 cycles of denaturation at 950C at 60 seconds, primer annealing at 560C for 75 seconds, and primer extension at 720C for 45 seconds. The PCR products were confirmed on a 1.5% agarose in 0.5% TBE buffer, visualized under ultraviolet light and eluted (Qiaquick gel extraction kit) for purification of the PCR product.
Ligation and transformation
The purified PCR products were ligated in pTZ57R/T vector using InsT/Aclone™ PCR product cloning kit (Cat no. K1213, MBI Fermentas). Ligation of the PCR fragment was confirmed by PCR using M13 specific primers targeting M13 sequences contained in pTZ57R vector. Ligated recombinant vector was transformed in competent E. coli (DH5-α) cells. Recombinant clones were selected by Blue-white screening. Recombinant plasmids were isolated and used for cycle sequencing, that were later processed for sequencing using the universal M13 primers.
Sequencing and SNP detection
Extension products obtained from cycle sequencing reaction were purified by vacuum manifold using AcroPrep™ 96 filter plates, (Cat. no. 5033, Omega 3K, Pall Corporation, USA). Cycle sequencing products were run on ABIPRISM automated DNA sequencer and raw data was collected by 310 Data Collection Software (Version 3.1.0). Sequences obtained by sequence analyzer were curated and vector sequences were removed using Vecscreen (NCBI) programme. These filtered sequences were then subjected to local alignment with Genebank database sequences using BLASTn protocol available at NCBI.
A consensus sequence of OLR1 Intron I of buffalo was generated and used as a reference sequence. Forty sequences were compared by multiple sequence alignment by using a multiple sequence alignment tool, Bioedit (v 7.0.7.1). SNP detection was done by multiple sequence alignment of consensus sequences of Bubalus bubalis in Bioedit (v 7.0.7.1) and MEGA4. SNP in a sequence was judged as a mismatch between samples.
Results and Discussion
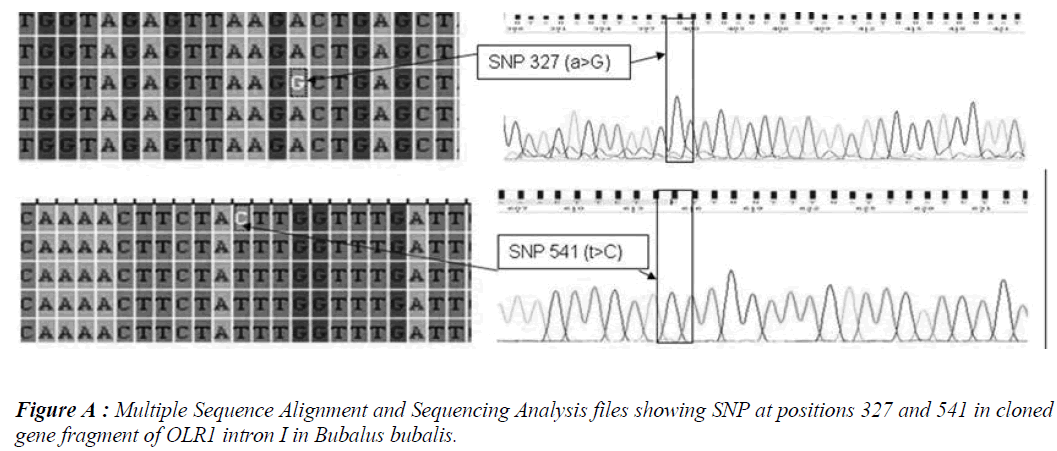
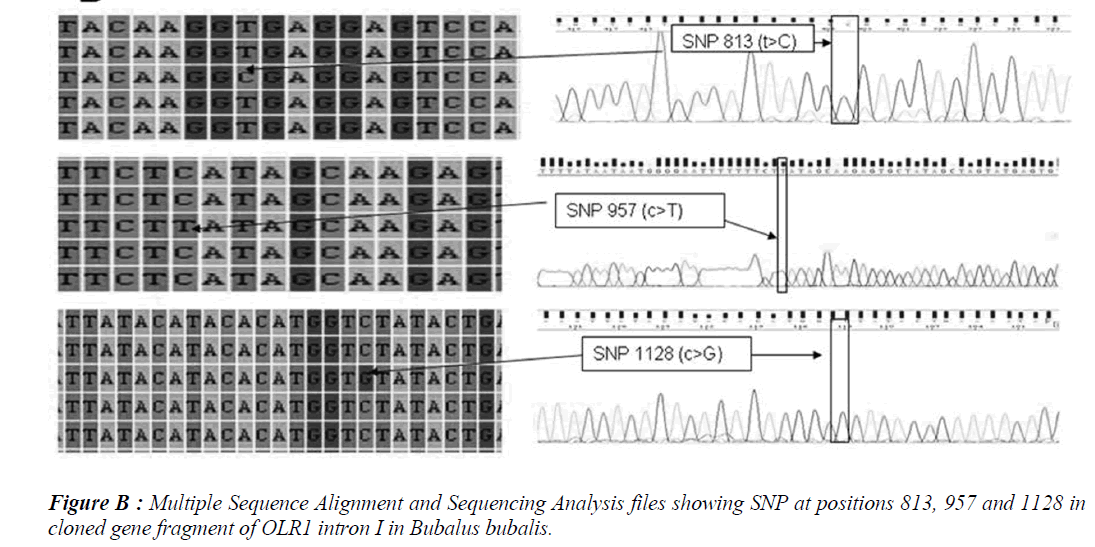
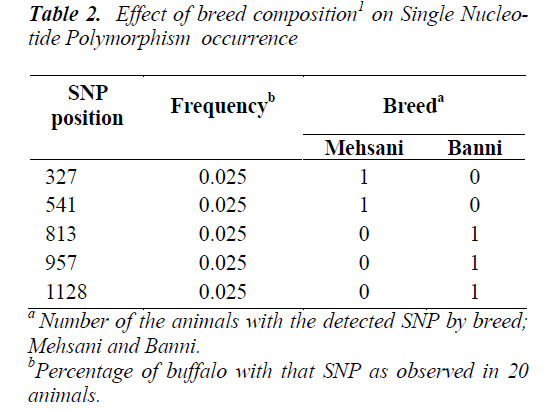
Five SNPs (Fig. 1 and Fig. 2) were detected within the 1163 bp fragment of OLR1 intron I gene in four breeds of Bubalus bubalis (Table 1). The frequency and breed composition of each of the six SNPs detected in the present study are summarized in the Table 2. The presence of all the SNP was affected by breed. The presence of SNP at nucleotide positions 327 and 541 were associated with the Mehsani breed where as SNPs at positions 813 and 957 found their association with Banni breed. SNP at all positions viz. 327, 541, 813, 846, 957 and 1128 were present in 2.5% of the population studied.
India possesses eleven recognized breeds of buffalo. Present study was conducted in two breeds found in Gujarat state. Mehsani and banni are the medium sized breeds, having a high milk fat percentage (>8 %). The most abundant type of variation in human and cattle genomes is the single nucleotide polymorphism or SNP, where a single base pair has been changed. Millions of SNPs have been found in humans, and there are over 600,000 in cattle with more being discovered every day [8]. In our earlier study we screened the same region of OLR1 in Jaffarabadi and Surti breeds of buffalo where in we explored three SNPs at nucleotide position 423 and 866 in surti buffalo and at 843 in Jaffarabadi buffalo. Various studies in bovine species have been conducted around the globe to derive an association of OLR1 gene polymorphisms with the milk fat percentage. In a study conducted in Italian brown swiss involving the genotyping for a SNP at position 8232 in OLR1 (NW_215807:g.8232C>A) revealed a frequency of 0.95 for the g.8232C allele [9]. In another study conducted in Dutch Holstein-Friesian cattle population in OLR 1 gene revealed that OLRg.8232C>A had a significant effect (P < 0.05) on milk-fat percentage [2]. A allele C of SNP 8232 in 3’ UTR in OLR1 gene had significant effects on fat yield and fat percentage [1]. Breed specific SNPs are not uncommon. In a study conducted on heat shock protein 70 in pure Brahman, pure Angus and their intercross breeds of cattle, breed specific SNPs were explored that tended to be associated with the particular breeds only [10]. The SNPs detected in the present study also followed breed lineage. All the SNPs showed breed specific occurrence which is fully in agreement with our previous study where in we explored three SNPs in Surti and Jaffarabadi breeds of Buffalo and all the three SNPs were found to be breed specific [11]. However, as these SNPs are present in intronic region, their functional significance is required to be verified. The OLR1 sequences and SNPs explored in Mehsana and banni breeds of buffalo are accessible in National Centre of Biotechnology Information (NCBI) and their Genbank accession numbers and SNP ID’s have been enlisted in the Table 1.
Acknowledgements
We are thankful to the Indian Council of Agricultural Research (ICAR) for providing us with the necessary funding for carrying out this research work
References
- Khatib H, Leonard SD, Schutzkus V, Luo W, Chang YM. Association of the OLR1 gene with milk compo- sition in hostein dairy cattle. J Dairy Sci 2006; 89: 1753-1760.
- Schennink A, Bovenhuis H, Léon-Kloosterziel KM, van Arendonk JAM, Visker MHPW.. Effect of poly- morphisms in the FASN, OLR1, PPARGC1A, PRL and STAT5A genes on bovine milk-fat composition. Ani Genet 2009; 40: 909-916
- Mehta JL, Li DY. Identification and autoregulation of receptor for OX-LDL in cultured human coronary ar- tery endothelial cells. Biochem Biophys Res Commun 1998; 248: 511-514.
- Imanishi T, Hano T, Sawamura T, Takarada S, Nishio Oxidized low density lipoprotein potentiation of Fas- induced apoptosis through lectin-like oxidized-low density lipoprotein receptor-1 in human umbilical vas- cular endothelial cells. Circ J 2002; 66: 1060-1064.
- Sawamura TN, Kume T, Aoyama H, Moriwaki H, Ho- shikawa Y, Aiba T, Tanaka S, Miwa Y, Katsura T, Kita, Masaki T. An endothelial receptor for oxidized low- density lipoprotein. Nature 1997; 386:73-77.
- Oka KT, Sawamura K, Kikuta S, Itokawa N, Kume T, Kita, Masaki T. Lectin-like oxidized low-density lipo- protein receptor 1 mediates phagocytosis of aged/ apoptotic cells in endothelial cells. Proc Natl Acad Sci 1998; 95: 9535-9540.
- Aoyama TT, Sawamura Y, Furutani R, Matsuoka MC, Yoshida H, Fujiwara, Masaki T. Structure and chromo- somal assignment of the human lectin-like oxidized low-density-lipoprotein receptor-1 (LOX-1) gene. Bio- chem J 1999; 339: 177-184.
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genet 2001; 157: 1819-1829.
- Khatib H, Rosa GJM, Weigel K, Schiavini F, Santus E, Bagnato A. Additional support for an association be- tween OLR1 and milk fat traits in cattle. Ani Genet 2007; 38: 308-310
- Lamb M, Okimoto R, Brown M, Rosenkrans C Jr, Asso- ciations Between Cattle Breed and Heat Shock Protein 70 Gene. AAES Res Series 2006; 545: 205-206.
- Shabir N, Jawale CV, Bhong CD, Kurkute AS, Patel TB, Rank DN, Joshi CG. SNP exploration in the Oxidised Low Density Lipoprotein Receptor 1 (OLR1) gene in Bubalus Bubalis. Buff Bulletin 2011; 30: 267-271.