Research Article - Journal of Systems Biology & Proteome Research (2017) Journal of Systems Biology & Proteome Research (Special Issue-2017)
Austroinulin from Stevia rebaudiana a Potential Agonist of the Peroxisome Proliferator-activated Receptor (PPAR?): an in-silico Study
Damilohun Metibemu1*, Akinloye OA3, Akinbamiwa BA1, Adeniran OY2, Eniafe OG1, Akamo AJ3, Omotuyi IO1
1Centre for Biocomputing and Drug Development, Adekunle Ajasin University, Akungba-Akoko, Ondo State, Nigeria
2Department of Biochemistry, Adekunle Ajasin University, Akungba-Akoko.
3Department of Biochemistry, Federal University of Agriculture, Abeokuta, Nigeria.
- Corresponding Author:
- Damilohun Metibemu
Centre for Biocomputing and Drug Development
Adekunle Ajasin University
Akungba-Akoko
Ondo State
Nigeria
E-mail: damilohun.metibemu@aaua.edu.ng.
Accepted Date: October 30, 2017
Citation: Metibemu D, Akinloye OA, Akinbamiwa BA, et al. Austroinulin from Stevia rebaudiana a Potential Agonist of the Peroxisome Proliferator-activated Receptor (PPARγ): an in-silico Study. J Syst Biol Proteome Res. 2017;1(1):10-15
Abstract
Activators (agonists) of the nuclear receptor, peroxisome proliferator-activated receptor (PPAR?) are therapeutically used to tackle hyperglycemia associated with type 2 diabetes. Current therapeutic compounds, thiazolidinedione used to sensitize cells to insulin have been reported to adversely affect the body. In view of this, the discovery of novel ligands is highly pertinent. Plant products have been documented, to be a promising pool of structures for drug discovery, and the PPAR?-activating potential of a wide range of natural products have been reported. In the present study, Computer-aided drug design techniques were employed in the screening of pharmacologically relevant compounds of medicinal plant. Seventy (70) phyto-ligands (phytochemicals) characterized from Stevia rebaudiana and obtained from literatures were screened for their agonistic effects on peroxisome proliferator-activated receptor (PPAR?) belonging to the nuclear receptor family. Austroinulin was the lead compound with a binding energy of -8.1kcal/mol. The pharmacological kinetic analysis of austroinulin was determined. The results of this study demonstrated that the phyto-ligand (austroinulin) exhibit agonistic activity on PPAR? and could be a potential insulin sensitizer
Keywords
Peroxisome proliferator-activated receptor (PPARγ); Thiazolidinediones; Stevia rebaudiana; Molecular docking; Austroinulin; Type 2 diabetes.
Introduction
Type II diabetes (T2D) is a chronic disorder that constitutes about 90–95% of diabetes mellitus (DM). It is a heterogeneous group of metabolic disorders characterized by an elevated blood glucose level. Type II diabetes [1] is recognized as one of the dominant executors of disability and casualty in the modern world. T2D is characterized by preliminary loss of cells' insulinsensitivity or a complementary deficiency in insulin secretion or both [2], resulting from the combination of both environmental factors and genetic composition [3].
WHO reported the global of diabetes mellitus (DM) [4] as of 1980 to be 108 million people, and over 110 million people in 1994 [5] and increased to 285 million people in 2010 [6], with a conceived future increment to 761 million by 2030 [7]. DM has been one of the top killing diseases for some decades, with an estimated value of 1.5-5.0 million deaths in 2012-2015 (IDF, 2006) owing to inefficient medical treatment.
Current therapeutic compounds used to sensitize cells to insulin, majorly the Thiazolidinediones, which when bind to PPARγ activate the transcription of genes (e.g., adipocyte fatty acid-binding protein, lipoprotein lipase, and phosphoenol pyruvate carboxykinase in adipose tissue) involved in glucose metabolism and maturation of adipocytes, and the up-regulation of acyl-Co synthetase, fatty acid transporters, and uncoupling protein-2 are recently reported to exacerbate other life-threatening side effects which lead to their limited usage [8,9]. Hence, the need for alternative therapeutic agents with little or no side effects [10].
In the present study, Seventy (70) phytoligands (phytochemicals) characterized from Stevia rebaudiana and obtained from literatures were screened for their agonistic effects on peroxisome proliferator-activated receptor (PPARγ) belonging to the nuclear receptor family. Stevia rebaudian is a perennial shrub of the family Asteraceae. This plant is reported to produce a diverse range of bioactive molecules that possess antibacterial, antifungal, antiprotozoal, and anti-diabetic properties [11]. The drug likeness of the lead phytochemical compounds were determine through the calculation of their molecular properties; calculated logP, polar surface area, number of hydrogen bond donor, number of hydrogen bond acceptors and molecular weight (Lipinski's rule) [12].
Materials and Methods
Preparation of phyto-ligands
Seventy (70) Phytochemicals that have been characterized from S. rebaudiana were obtained from literatures. The structures of some of the phytochemicals were drawn with the ChemAxon software; MarvinSketch v15.11.30 and saved in the sdf format, while the other structures of the phytochemicals were downloaded in the sdf format from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). All phytochemicals were converted from their sdf format to the pdb format by Open Babel and finally to the pdbqt using ligprep command lines.
Preparation of standard
The standard drugs used in the present study, Pioglitazone and Rosiglitazone, are a group of synthetic drugs named Thiazolidinediones. Pioglitazone and Rosiglitazone are the two potent well-known drugs of the class Thiazolidinediones that have been successfully used in restoring cells' sensitivity to insulin, though banned in some quarters due to its reported life threatening side effects [13-14]. The structure of the standards (Pioglitazone and Rosiglitazone) were obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov)
Preparation of target protein
The Crystal structure of the ligand-binding domain of the human PPAR-gamma in complex with an agonist with crystallographic resolution of 2.07Aº with PDB ID: 2F4D was downloaded from the protein data bank (http://www.rcsb.org). PyMOL plugin in Autodock/Vina was employed to extract the receptor. The SWISS-MODEL software was used to rebuild the missing residues and atoms of the human PPAR-gamma.
Molecular docking and virtual high throughput screening
Virtual high throughput screening (vHTS) method was employed, to screen a pool of the compounds library to investigate the binding affinity cum agonistic properties of the phytochemicals from Stevia rebaudiana against the binding pocket of the human PPAR-gamma target receptor [15]. To ensure the phytochemicals occupy the exact binding pocket as obtained in the co-crystallized compound, the grid coordinates was set exactly as those of the co-crystallized compound; x= 9.24, y= -5.89, z= 39.09. Force fields were applied to the phytochemicals for subsequent conversion to 3D and later to pdbqt, using command lines in auto dock vina, and then, processed for docking.
Validation of docking results
The docking results were validated with the blasting of the fasta sequences of the crystal structure of the ligand-binding domain of the human PPAR-gamma (ID: 2F4D), obtained from the protein data bank unto the online available ChEMBL Database (www.ebi.ac.uk/chembl/). The bioactivity generated by the database, having an identity of 100%, EC50 value of 2599, and KI value of 312, was downloaded in txt format. The bioactivity was sorted out; missing or misplaced data were removed. Only 88 of the total 2599 drug-like compounds were recovered. The compiled compounds were split and converted to 2D (in sdf format) by Data Warrior software (version 2), converted to pdb format by Open Babel, and finally to pdbqt. The ligands were docked into the binding domain of PPARγ (as indicated by the config file). The config file indicated the grid map of the active site of PPAR-gamma generated by AudoDock Vina. A correlation coefficient graph was plotted between the docking scores of the 88 compounds generated and their corresponding pIC50 (experimentally determined) values. Spearman Rank correlation co-efficient graph was plotted to obtain the correlation (R²) between the dockings results of the ChEMBl’s compounds and their corresponding experimentally generated results.
Result and Discussion
Other than the testing of extracts and bio-guided approaches, virtual screening remained an effective approach for the discovery of novel PPARγ ligands from natural sources [16]. Numerous 2D and 3D virtual screening techniques have successfully provided structurally diverse natural product PPARγ activators, hence, revealing natural products as a rich source for novel PPARγ agonists [16].
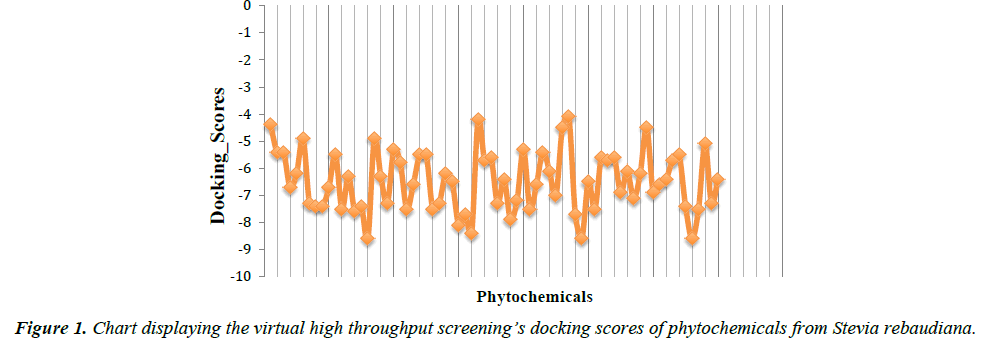
In the present study, virtual high throughput screening (vHTS) technique was employed to screen a pool of seventy (70) phytocompounds from Stevia rebaudiana plant for their PPARγ agonistic properties. Austroinulin was discovered as the lead compound with the binding energy of -8.1 kcal/mol (appendix). The drug-likeness of austroinulin was confirmed by subjecting it to the Lipinski’s rule of five, interestingly the lead compound, austroinulin violated none of the rules and with three (3) number of rotatable bonds, it possesses good bioavailability (number of rotatable bonds influences the bioavailability and the binding potency) (Tables 1 and 2).
| Compound Name | Number of rotatable bonds | Binding Energy | |
|---|---|---|---|
| (Kcal/mol) | |||
| Austroinulin | 3 | -8.1 | |
| Thiazolidinediones | Pioglitazone | 7 | -8.5 |
| Rosiglitazone | 7 | -7.1 | |
Table 1: Showing the number of rotatable bonds in Austroinulin and in the standard drugs (Pioglitazone and Rosiglitazone). Compounds that meet the criteria of (1) 10 or fewer rotatable bonds and (2) polar surface area ≤140 Å2 (or 12 or fewer H-bond donors and acceptors) have high probability of good oral bioavailability. The number of rotatable bonds influences bioavailability and binding potency.
| Molecular Properties | Lipinski's Rule Of Five | Austroinulin Drug-Like Properties |
|---|---|---|
| Molecular Mass | <500 Dalton | 322.48216 |
| Hydrogen Bond Acceptors | <10 | 3 |
| Hydrogen Bond Donors | <5 | 3 |
| High Lipophilicity | < 5 | 4.5 |
| (Log P) | ||
| Molar Refractivity | Between 40-130 | 94.87 |
| Polar Surface area | ≤ 140 Å2 | 60.69 |
Table 2: Lipinski's drug-like properties of austroinulin: The rule describes drug candidate’s pharmacokinetics in the human body which also including their absorption, distribution, metabolism, and excretion (“ADME”). Austroinulin, violated none of the rules. This table was generated using the ChemAxon suite.
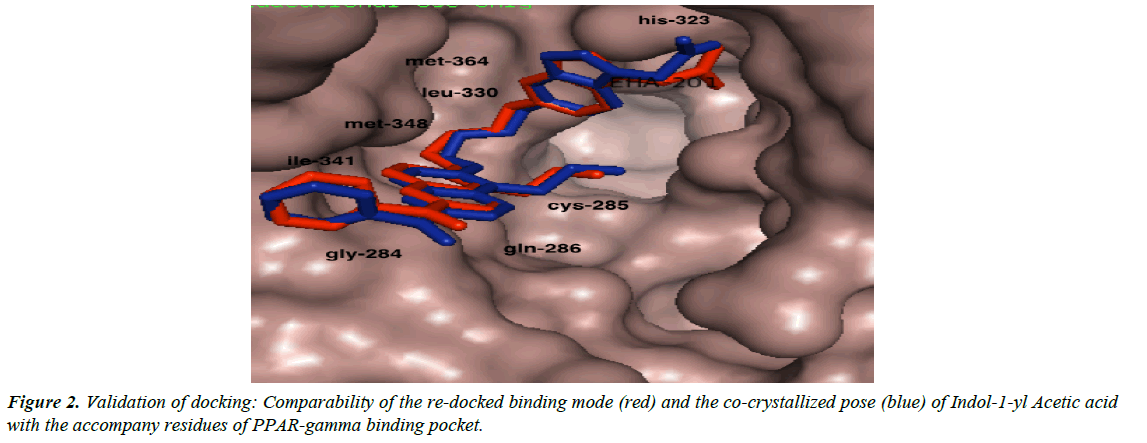
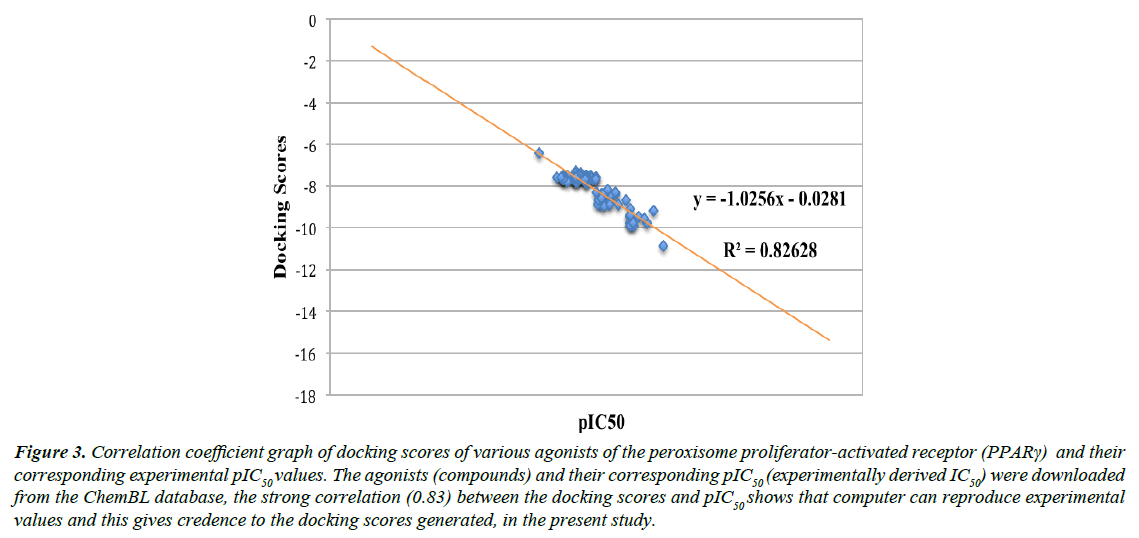
We validated the accuracy of our docking protocol by redocking the co-crystallized ligand (Indol-1-yl Acetic acid) back into the binding pocket of the PPARγ (2F4B1) [17], (Figure 1). As revealed above, the re-docked pose overlapped almost totally with the experimental orientation, indicating that autodock vina with a very high accuracy re-docked the co-crystallized Indol- 1-yl Acetic acid back into the binding pocket of the PPARγ, this implies that our docking methodology was reliable and the docking scores obtained are correct. The accuracy of our docking scores was further validated using the online available ChEMBL Database, the Fasta sequence of the crystal structure of the Human PPAR-gamma (ID: 2F4B) was blasted on www. ebi.ac.uk/chembl/. The compounds obtained were docked into the ligand-binding domain of the Human PPAR-gamma, a correlation coefficient graph plotted between the docking scores of the compounds generated and their corresponding ChEMBL’s Pchem values (experimentally determined IC 50), Figure 2. The strong correlation coefficient between the docking scores and the experimentally derived data in the present study, lend credence to the fact that computational experiment can replicate experimental data at least in this present study and that our docking scores (appendix) using AutoDock Vina algorithm is dependable (Figure 3).
Figure 3: Correlation coefficient graph of docking scores of various agonists of the peroxisome proliferator-activated receptor (PPARγ) and their corresponding experimental pIC50 values. The agonists (compounds) and their corresponding pIC50 (experimentally derived IC50) were downloaded from the ChemBL database, the strong correlation (0.83) between the docking scores and pIC50 shows that computer can reproduce experimental values and this gives credence to the docking scores generated, in the present study.
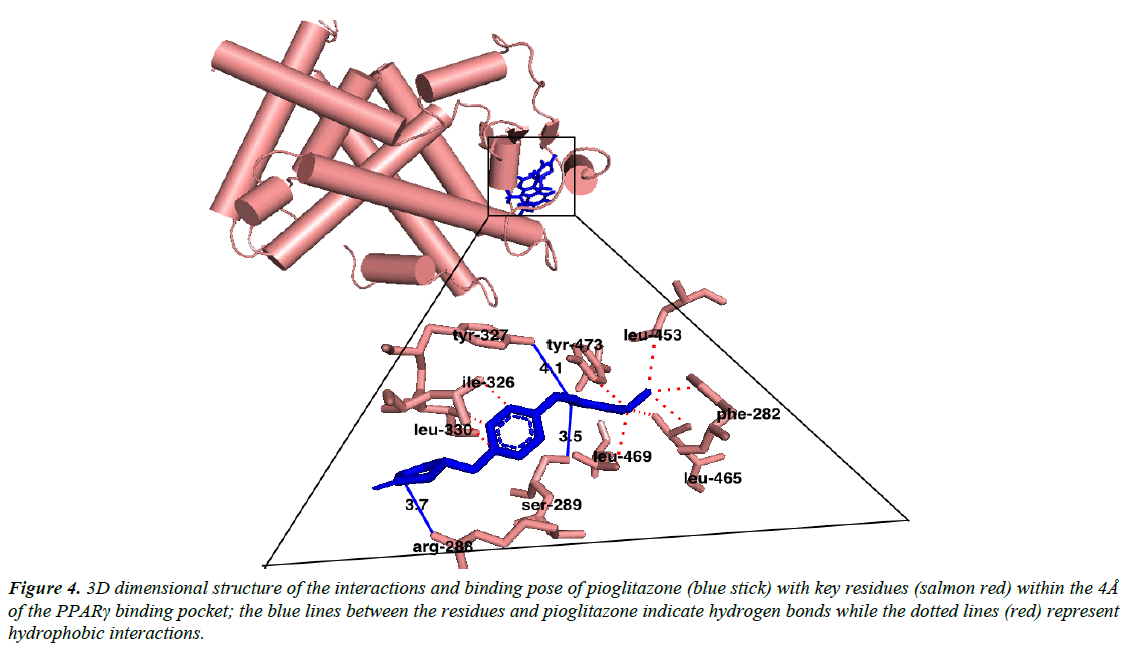
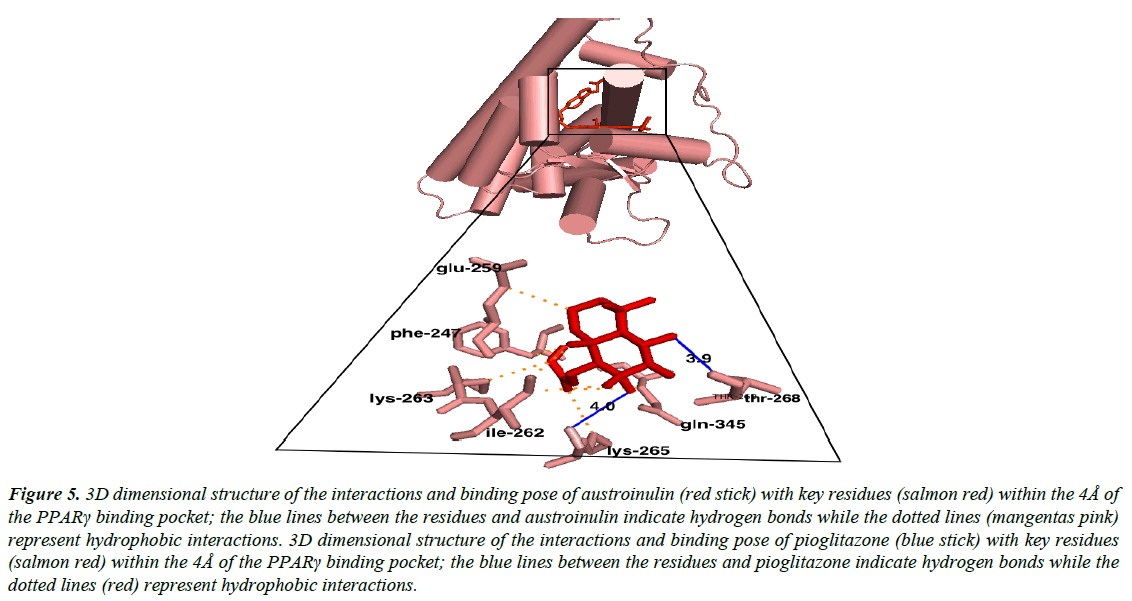
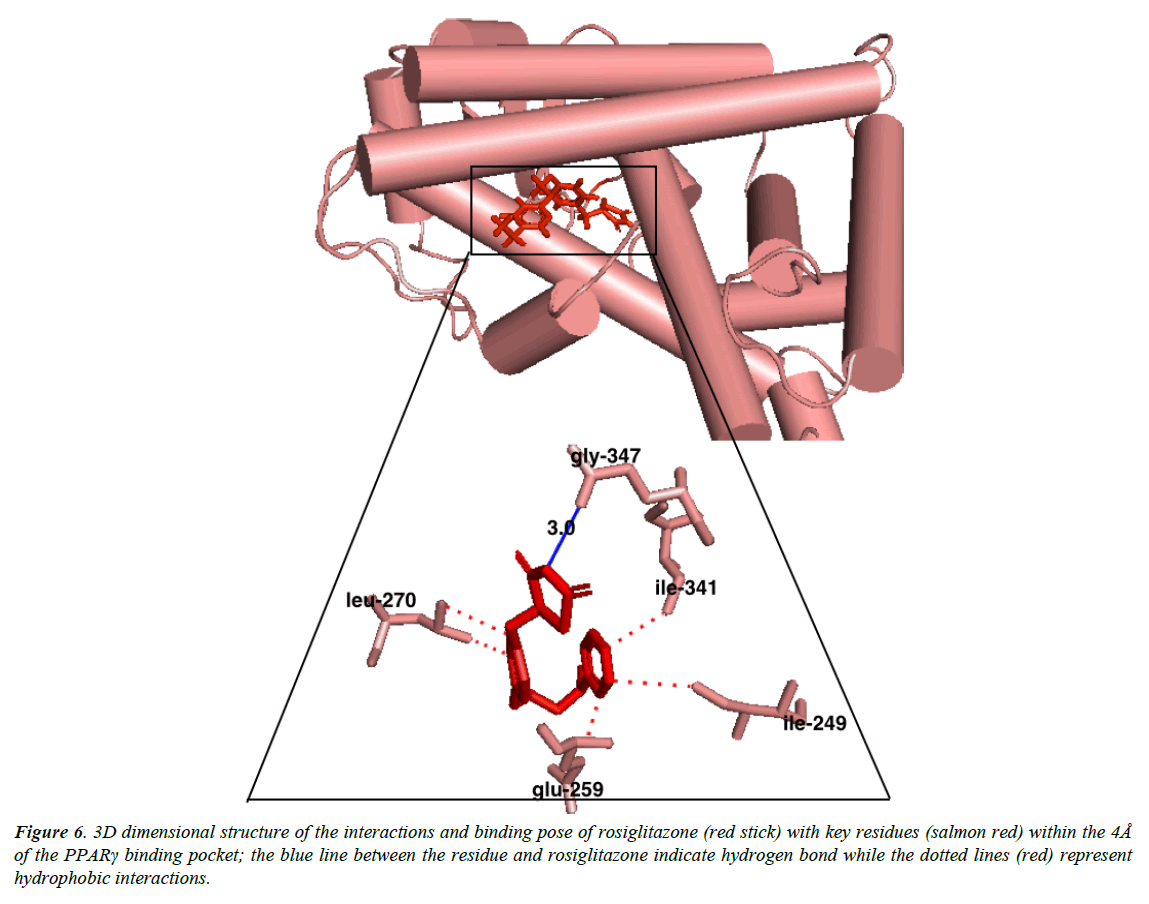
Austroinulin, the lead compound has a binding energy of -8.1 kcal/mol, while the standard drugs pioglitazone and rosiglitazone have binding energies of -8.5 kcal/mol and -7.1 kcal/mol respectively (Table 3). The highest binding energy (-8.5kcal/ mol) accrued to pioglitazone in this regard is believed to be due to the extensive high number of hydrophobic interactions (sixteen) of pioglitazone with key residues at the active site of the PPARγ; Met – 329, Ala-2923, Ile-326, Leu-336, Phe-282, His-323, Leu-453, Leu-469, His-449, Leu-465, Gln-286, Cys- 285, Leu-333 (Figure 4) and coupled with the high number of hydrogen bonds (Arg-288, Tyr-474 and Ser-289). However, Autstroinulin and rosiglitazone though possess different binding energies (-8.1 kcal/mol, -7.1 kcal/mol respectively) shared some common amino acids interactions (six) at the binding pocket of PPARγ (Figure 5 and 6); Leu-270, ser-342, Ile-262, Gln-345, Gly-258 and Glu-259.
| S/N | Phytochemicals | Docking scores |
|---|---|---|
| 1 | Austroinulin | -8.1 |
| 2 | Caryophyllene_Oxide | -8 |
| 3 | Jhanol | -8 |
| 4 | Steviol | -8 |
| 5 | Catechin | 7.9 |
| 6 | Cyclohexadecane | -7.9 |
Table 3: Showing the docking scores of the six (6) lead phytochemicals from Stevia rebaudiana.
Figure 4: 3D dimensional structure of the interactions and binding pose of pioglitazone (blue stick) with key residues (salmon red) within the 4Å of the PPARγ binding pocket; the blue lines between the residues and pioglitazone indicate hydrogen bonds while the dotted lines (red) represent hydrophobic interactions.
Figure 5: 3D dimensional structure of the interactions and binding pose of austroinulin (red stick) with key residues (salmon red) within the 4Å of the PPARγ binding pocket; the blue lines between the residues and austroinulin indicate hydrogen bonds while the dotted lines (mangentas pink) represent hydrophobic interactions. 3D dimensional structure of the interactions and binding pose of pioglitazone (blue stick) with key residues (salmon red) within the 4Å of the PPARγ binding pocket; the blue lines between the residues and pioglitazone indicate hydrogen bonds while the dotted lines (red) represent hydrophobic interactions.
Autroinulin has a higher binding energy (-8.1 kcal/mol) when compared with rosiglitazone (-7.1 kcal/mol) (Tabe 1), there are a total of eleven amino acids interaction of austroinulin with the binding pocket of PPARγ (Figure. 4); Leu-270, Ser- 342, Cys-261, Gly-346, Phe-247, Gln-345, Gly-258, Glu-259, Thr-268 ( Ten (10) hydrophobic interactions and two (2) hydrogen bonds interaction; Thr-268 and Lys-265) (Figure 4), also, rosiglitazone formed a total of eleven (11) interactions within the 4Å of the PPARγ binding pocket (Figure. 6) ; Lys-265, Gly-347, Ile- 341, Ser- 342, Ile-249, Ile-281, Arg-280, Glu-259, Leu-270, Ile-262, Gly-258 ( Ten (10) hydrophobic interactions and one (1) hydrogen bond interaction – Gly-347). The differences in the binding energies of pioglitazone, austroinulin (the lead compound) and rosiglitazone is believed to be as a result of the variations in the amino acids interactions most likely the hydrogen bond interactions of the three compounds with critical residues within the binding pocket of the PPARγ; Pioglitazone with three (3) hydrogen bonds, austrinulin with two (2) hydrogen bonds and pioglitazone with only one (1) hydrogen bond.
Figure 6: 3D dimensional structure of the interactions and binding pose of rosiglitazone (red stick) with key residues (salmon red) within the 4Å of the PPARγ binding pocket; the blue line between the residue and rosiglitazone indicate hydrogen bond while the dotted lines (red) represent hydrophobic interactions.
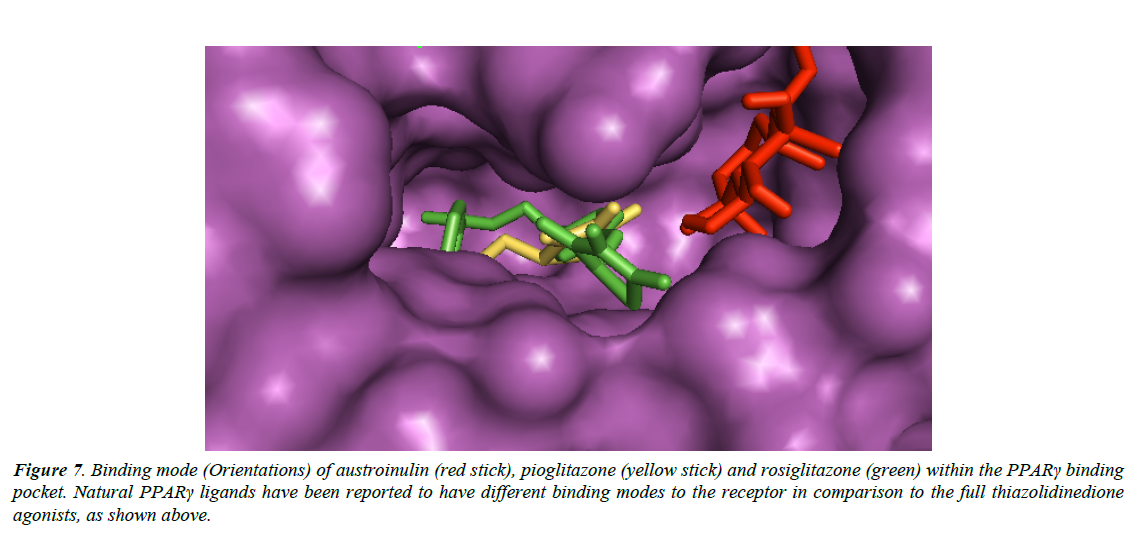
Natural PPARγ agonists have different binding modes (Figure 7) when compared with the thiazolidinedione agonists, and these may be associated with the reduced side effects of natural agonists of PPARγ (Wang et al., 2014).
Figure 7: Binding mode (Orientations) of austroinulin (red stick), pioglitazone (yellow stick) and rosiglitazone (green) within the PPARγ binding pocket. Natural PPARγ ligands have been reported to have different binding modes to the receptor in comparison to the full thiazolidinedione agonists, as shown above.
Conclusions
There have been few or little reports in the literatures, on the active constituents of numerous medicinal plants extracts indicated to be activators of the PPARγ. Austoinulin from Stevia rebaudiana, bear a good potential as a possible agonist of the PPARγ, hence, it needs to be further investigated both in-vitro and in-vivo.
References
- International Diabetes Federation. IDF. Diabetes Atlas (7th edn.) Brussels, Belgium (2006).
- Metibemu DS, Saliu JA, Metibemu AO et al. Molecular Docking Studies of Isorhamnetin from Corchorus olitorius with Target Alpha-amylase Related to Type 2 Diabetes. 2016. J Chem Pharm Res. 8(4): 1262-66.
- Riserus U, Willett W, Hu F. Dietary fats and prevention of type 2 diabetes. Prog. Lipid Res. 2009; 48(1):44–51.
- Libby G, Donnelly LA, Donnan PT, et al. New users of metformin are at low risk of incident cancer. Diabetes Care. 2009; 32(9):1620-5.
- McCarty D, Zimmet P. Diabetes 1994 to 2010: Global estimates and projections. Melbourne: International Diabetes Institute 1994.
- Williams, Textbook of endocrinology (12th ed.). Philadelphia, 2013. pp. 1371-1435
- Wild S, Roglic G, Green A, et al. Global Prevalence of Diabetes: Estimates for the year 2000 and projections for 2030. Diabetes Care. 2004; 27(5):1047–53
- Forman LM, Simmons DA, Diamond RH. Hepatic failure in a patient taking rosiglitazone. Ann. Intern. Med. 2000;132(2):118
- Willson TM, Brown PJ, Sternbach DD, et al. The PPARs: From orphan receptors to drug discovery. J. Med. Chem. 2000;43(4):527–50
- Kameswararao RB, Kesavulu MM, Apparao C. Fitoterapia. 2003:74:7-13.
- Brandle JE, Starratt AN, Gijzen M. Stevia rebaudiana: Its agricultural, biological, and chemical properties. CAN J PLANT SCI. 1998;78(4):527–36
- Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today. 2004;1(4):337–41
- Diamond GA, Kaul S. Rosiglitazone and cardiovascular risk. N Engl J Med. 2007; 357(9):937–40
- Singh S, Loke YK, Furberg CD. Long-term risk of cardiovascular events with rosiglitazone. JAMA 2007; 298(10):1189
- Shoichet BK. Virtual screening of chemical libraries. Nature. 2004; 432(7019):862
- Wang L, Waltenberger B, Pferschy-Wenzig EM, et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): A review. Biochemical pharmacology. 2014; 92(1):73-89.
- Morris GM, Goodsell DS, Halliday RS, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of computational chemistry. 1998; 19(14):1639-62.