Review Article - Journal of Systems Biology & Proteome Research (2017) Journal of Systems Biology & Proteome Research (Special Issue-2017)
Applications of Targeted Proteomics and Mass Spectrometry in Trastuzumab Pharmacokinetics Assessments
Rajasekhar Baru*
Granada Pharma Ltd., Granada, Riyadh 3939, Saudi Arabia
- Corresponding Author:
- Dr. Rajasekhar Baru
Granada Pharma Ltd.
Granada
Riyadh 3939 Saudi Arabia
Tel: + 966-583956618
E-mail: barurajasekhar@gmail.com
Accepted Date: October 09, 2017
Citation: Baru R. Applications of Targeted Proteomics and Mass Spectrometry in Trastuzumab Pharmacokinetics Assessments. J Syst Biol Proteome Res. 2017;1(1):7-9
Abstract
The therapeutic monoclonal antibody drug Trastuzumab is widely used for treating metastastatic breast cancer patients with overexpression of HER2 on the tumor. Quantitative targeted proteomics based approaches utilize LC-MS technologies and are evolving as a complementary technique to standard ligand-binding based assays. In the current review, a surrogate peptide based quantitative proteomics assessment by selecting specific signature peptides from the complementary determining region of Trastuzumab is discussed. Double stable isotope labelling method utilizing two surrogate peptides to evaluate accurate quantification of the target analyte peptide of Trastuzumab is discussed. This mini-review highlights the strength of the double stable isotope label approach for the quantitative evaluation of monoclonal antibody biologics in a human biological matrix.
Introduction
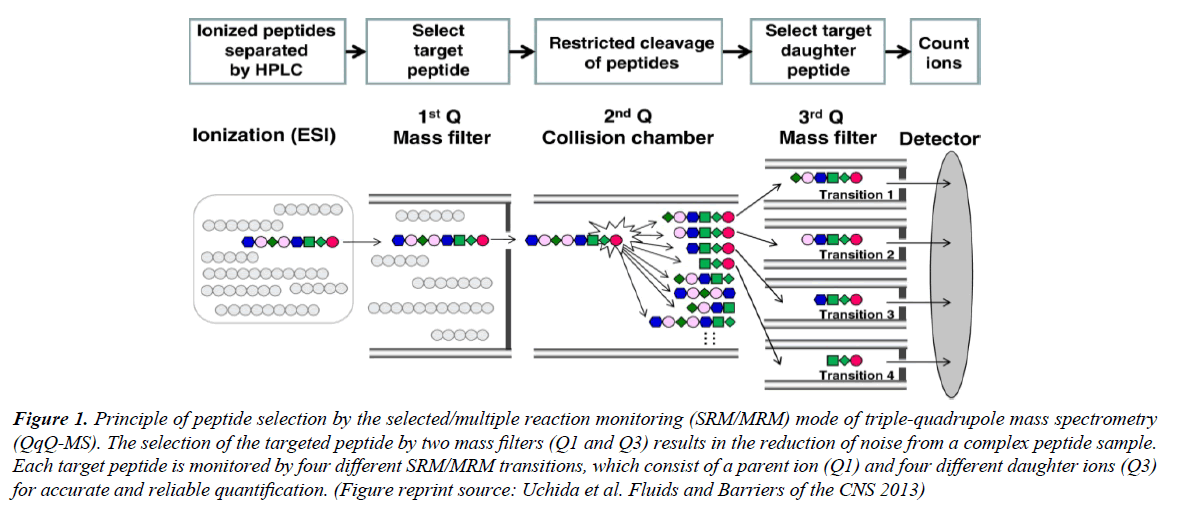
Targeted proteomics has emerged as a powerful protein quantification tool making a strong impact in systems biology and biomedical research, deployment of this technology in clinical applications is on the horizon. Revolutionary improvements in mass spectrometry (MS) technology have resulted in state-of-the-art MS platforms with exquisite selectivity and sensitivity levels for achieving accurate quantification. A triplequadrupole mass spectrometer (QqQ-MS) consists of three single-quadrupole mass analyzers and serves as the central core technology coupled to liquid chromatography (LC) in targeted proteomics studies. The Q1 section of a mass spectrometer acts as a mass filter that enables the selection of ‘precursor ions’. Peptide fragmentation occurs in the Q2 section by collisioninduced dissociation. Finally, the fragmented ‘product ions’ are selected in Q3 (Figure 1) and are further guided to the detector for quantification, resulting in a trace plot of signal intensity versus retention time for each precursor ion-product ion pair. LC-MS/ MS is accurate, precise, and enables throughput analysis, and thus several LC-MS/MS studies have been dedicated to the absolute quantification of intact proteins in biological fluids [1–3].
Figure 1: Principle of peptide selection by the selected/multiple reaction monitoring (SRM/MRM) mode of triple-quadrupole mass spectrometry (QqQ-MS). The selection of the targeted peptide by two mass filters (Q1 and Q3) results in the reduction of noise from a complex peptide sample. Each target peptide is monitored by four different SRM/MRM transitions, which consist of a parent ion (Q1) and four different daughter ions (Q3) for accurate and reliable quantification. (Figure reprint source: Uchida et al. Fluids and Barriers of the CNS 2013)
Clinical applications of targeted proteomics mainly include biomarker evaluation; however, the field of monoclonal antibody (mAb)-dosed pharmacokinetics (PK) has recently begun to embrace the strength of LC-MS as a complementary tool to ligand-binding assays (LBAs). The goal of this report is to highlight the developments of targeted proteomics-based assays in PK assessments of mAbs, including trastuzumab. Trastuzumab is a recombinant immunoglobulin G1 (IgG1)- kappa humanized mAb that explicitly targets the extracellular juxtamembrane domain of HER2 (Human epidermal growth factor receptor 2) receptor, and is a frontline therapy for breast cancer patients with HER2 over expression levels improving overall survival rates for patients. High affinity binding of trastuzumab to its cognate epitope potentially inhibits the dimerization of HER2 receptor, inhibition of DNA repair, cellcycle arrest, antibody-dependent cellular cytotoxicity, and is commonly used in HER2-positive breast cancer treatment [4-6]. Market authorization approval of Trastuzumab (HERCLON™, Roche) was given by the US Food & Drug Administration in September 1998 and by the European Medicines Agency in August 2000 for the treatment of human metastatic breast cancer [7] and has become a promising treatment therapy. Trastuzumab is administered by intravenous infusions of 10 to 500 mg once every week with an initial dose of 8 mg/kg infused over 90 minutes, followed by 6 mg/kg over 30 to 90 minutes every 3 weeks. The compound has demonstrated dose-dependent PK, with an average half-life of 2 and 12 days at the 10 and 500 mg dose levels, respectively [8,9]. At the highest weekly dose studied (500 mg), the mean peak serum concentration was 377 μg/mL [10].
PK refers to the biological processes determining the absorption, distribution, metabolism, and excretion of a drug in an organism. During early mAb drug development, the PK characteristics of multiple candidates are among the critical considerations for lead candidate selection. PK studies of innovators and bio similar products at preclinical and clinical stages are critically evaluated by global drug approval regulatory authorities. PK assessments of therapeutic mAbs in biological fluids are widely performed using LBAs (Ligand binding assays) such as enzyme-linked immunosorbent assay (ELISA). However, there is a significant limitation to these assays, in that therapeutic mAbs can become analogs of endogenous IgGs in plasma, resulting in a minor change to their amino acid or nucleotide sequence by splicing variants, and the standard ELISA is unable to differentiate between endogenous and exogenous variants [11]. In addition, the specificity, accuracy, and reproducibility of the LBAs are inhibited by the presence of anti-mAbs. Cross reactivityrelated issues, the long time required for antibody development and matrix effects have all impeded the swift and successful utilization of this approach [12,13]. Alternatively, targeted proteomics and MS-based methodologies are available for accurately quantifying mAbs, offering superior selectivity over an immunoassay, and the development method is significantly shorter and less expensive. However, application of this approach is not straightforward for large molecules such as mAbs, and requires additional steps in sample preparation, including immunoaffinity depletion, enzymatic digestion, and quantification using a stable isotope-labeled internal standard surrogate peptide. Several preclinical studies have demonstrated the applicability of adopting an LC/MS approach as a surrogate to ELISA [14,15].
In this report, we do not provide a comprehensive assessment of the various clinical targeted proteomics studies conducted to date, but rather provide an overview of the targeted proteomicsbased methodologies used in PK assessments of trastuzumab. Quantification of trastuzumab in human plasma [16] and rat plasma [17] using the selected reaction monitoring (SRM) mode of LC-MS/MS and LC-time-of-flight-MS/MS, respectively; demonstrates target proteomics advancements in clinical studies.
Quantitative Targeted Proteomics and Skyline Multiple Reaction Monitoring (MRM) Transitions
Targeted proteomics is a focused-driven strategy, which has emerged as a powerful assay that is capable of selective and sensitive detection, as well as quantification of potentially any protein of interest in the complex proteome. The primary aim is to monitor a few selected proteins/peptides (signature peptides) of interest with high sensitivity, reproducibility, and quantitative accuracy using a QqQ-MS. The SRM/MRM process employing a QqQ-MS has higher sensitivity specifically towards low-abundant peptide fragments and higher quantitative precision compared to other MS methods. The Skyline Targeted Proteomics Environment [18] is an application for building quantitative MRM and full-scan methods for targeted proteomics.
Trastuzumab: Signature Peptide Selection
Signature peptide refers to an enzymatically cleaved peptide with a unique sequence that essentially originates from the complementary determining region (CDR) of the FaB segment of the mAb. Critical evaluation of the signature peptide is analyzed using various bioinformatics tools, including pBlast and the ClustalW algorithm based on GENETYX software (GENETYX, Tokyo, Japan). Trastuzumab MRM transitions with specific peptide sequences and charge states +2, FTISADTSK (504.772/667.341) and IYPTNGYTR (542.775/808.395) were evaluated with Skyline. Meticulous manual data analysis needs to be performed to filter out the peptides resulting from the fragment crystallizable (Fc) region of mAb and endogenous IgG’s in human serum possess high degree of sequence similarity that can potentially interfere with accurate quantification of the targeted mAb.
Double Stable Isotope Label (dSIL) Approach
The dSIL approach is used to quantify the therapeutic mAb trastuzumab in a complex serum matrix. This approach utilizes two unique surrogate tryptic peptides for evaluating the overall proteolytic digestion efficiency and accurate quantification of the target analyte peptide. Enzymatic digestion of a complex mAb produces several peptides, and thus a wide range of enzymes, including LysC, trypsin, GluC and pepsin, are used in proteomics studies. Trypsin is a preferred protease and it cleaves with high specificity targeting the C-terminal of basic amino acid residues (lysine and arginine) and the resulting tryptic peptide fragments achieve multiple charge states during ESI process and are amenable for MS/MS fragmentation and subsequent identification. A surrogate labeled peptide (SIL-IS) possesses intrinsic physical and chemical properties, identical separation efficiency on the chromatographic scale, correcting for variability and delivers enhanced quantitative precision in mass spectrometry analysis. This approach is widely used for the quantification of protein biotherapeutics, including mAbs and the unique selectivity of QqQ-MS differentiates the labeled isotopes of the same element of endogenous and surrogate labeled peptide. It mitigates the risks associated with background signal interference caused by complex biological matrices. The surrogate peptide approach mitigates the challenges involved in developing specific antibodies and is becoming a method choice of the quantification of large molecules.
Surrogate peptide-based quantitative proteomics assessment of trastuzumab (Herclon®, Roche) is performed by selecting specific signature peptides (FTISADTSK, IYPTNGYTR) from the CDR. Employing a uniformly heavy isotope-labeled common whole-mAb internal standard and a common immunocapture methodology for a therapeutic mAb was developed for a preclinical study [19].
Conclusions
This mini-review highlights the strength of the dSIL approach for the quantitative evaluation of mAb biologics in a human biological matrix. An LC-MS/MS-based targeted proteomics approach was applied for method development and method validation for evaluations of the PK of trastuzumab in human serum. We successfully developed a dSIL method for the precise and accurate quantification of trastuzumab ranging from 5 μg/ mL to 500 μg/mL. Validation experiments, including precision and accuracy batches, cross-contamination, autosampler carryover, and selectivity were performed as per regulatory guidelines. Validation experiments met the regulatory acceptance criteria, indicating the validation acceptability of the established method. The developed dSIL approach is potentially valuable for the evaluation of the PK of trastuzumab in clinical human serum samples. We are currently extending the method for the evaluation of a clinical study required for regulatory submission.
References
- Becher F, Pruvost A, Clement G, et al. Quantification of small therapeutic proteins in plasma by liquid chromatography− tandem mass spectrometry: Application to an Elastase Inhibitor EPI-hNE4. Anal. Chem. 2006; 78(7):2306-13.
- Budhraja RH, Shah MA, Suthar M, et al. LC-MS/MS Validation Analysis of trastuzumab using dSIL approach for evaluating pharmacokinetics. MOLECULES. 2016; 21(11):1464.
- Ji QC, Rodila R, Gage EM.et al. A strategy of plasma protein quantitation by selective reaction monitoring of an intact protein. Anal. Chem. 2003, 75, 7008–14.
- Slamon, DJ, Godolphin WJ, Holt J A. et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science 1989, 244, 707–12.
- Slamon D, Clark G, Wong S, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235(4785):177–82.
- Molina MA, Codony SJ, Albanell J, et al. Trastuzumab (Herclon), a humanized anti-Her2 receptor monoclonal antibody, inhibits basal and activated Her2 ectodomain cleavage in breast cancer cells. Cancer Res. 2001, 61, 4744–49
- Siegel, J.P. Biologics license application for trastuzumab. 1998.
- Damen CN, de Groot ER, Heij M, et al. Development and validation of an enzyme-linked immunosorbent assay for the quantification of trastuzumab in human serum and plasma. Anal. Biochem. 2009; 391(2):114–20.
- Sanchez AB, Nguyen T, Dema-Ala R, et al. A general process for the development of peptide-based immunoassays for monoclonal antibodies. Cancer Chemother. Pharmacol 2010; 66(5):919–25.
- Fusaro VA. Mani DR. Mesirov JP, Et al. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 2009, 27, 190–8
- Iwamoto N, Takanashi M, Hamada A, et al. Validated LC/MS bioanalysis of Rituximab CDR peptides using nano-surface and molecular-orientation limited (nSMOL) proteolysis. Biol. Pharm. Bull. 2016, 39, 1187–94.
- Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J. Immunol. Methods. 2009; 347(1):3-11.
- Ezan E, Bitsch F. Critical comparison of MS and immunoassays for the bioanalysis of therapeutic antibodies. Bioanalysis 2009; 1(8):1375–88.
- Heudi O, Barteau S, Zimmer D, et al. Towards absolute quantification of therapeutic monoclonal antibody in serum by LC− MS/MS using isotope-labeled antibody standard and protein cleavage isotope dilution mass spectrometry. Anal. Chem. 2008 May 9; 80(11):4200-7.
- Jenkins R, Duggan JX, Aubry AF, et al. Recommendations for validation of LC-MS/MS bioanalytical methods for protein biotherapeutics. The AAPS journal. 2015; 17(1):1-6.
- Hart MH, de Vrieze H, Wouters D, et al. Differential effect of drug interference in immunogenicity assays. J. Immunol. Methods. 2011; 372(1):196-203.
- Barnidge DR, Hall GD, Stocker JL, et al. Evaluation of a cleavable stable isotope labeled synthetic peptide for absolute protein quantification using LC− MS/MS. J Proteome Res. 2004; 3(3):658-61.
- MacLean B, Tomazela DM, Shulman N, et al. An open source document editor for creating and analyzing targeted proteomics experiments. BIOINFORMATICS. 2010; 26(7):966-8.
- Li H, Ortiz R, Tran L, et al. General LC-MS/MS method approach to quantify therapeutic monoclonal antibodies using a common whole antibody internal standard with application to preclinical studies. Analytical chemistry. 2012; 84(3):1267-73.