Research Article - Biomedical Research (2017) Volume 28, Issue 9
A new method of automatic recognition for tuberculosis disease diagnosis using support vector machines
Amani Yahiaoui1, Orhan Er2*, Nejat Yumusak11Department of Computer Engineering, Sakarya University, Adapazari, Turkey
2Department of Electrical and Electronics, Bozok University, Yozgat, Turkey
- *Corresponding Author:
- Orhan Er
Bozok Universitesi, Electrical - Electronics Engineering
Erdoğan Akdağ Kampus, Yozgat, 66200, Turkey
Accepted date: Febraury 28, 2017
Abstract
Tuberculosis is an infectious disease caused by a bacillus called Mycobacterium tuberculosis. It can lead to death in untreated and inappropriately treated patients particularly in countries with low income. Therefore, early diagnosis of the disease not only increases treatment success, but also reduces death rates. Today, due to high classification and diagnosis rates, specialist systems have become an important tool in diagnosis of the disease. In this study, support vector machines (SVM), which is a machine learning technique was used for preliminary diagnosis of tuberculosis disease for the first time. A recognition system that was developed with the properties included in patient reports obtained from a local hospital was tested for its performance. The results indicated performance of the designed system was quite successful and that it could be used in diagnosis of the disease. Obtained diagnostic results were compared with similar studies using different specialist systems on this disease, and it was observed that our results were better.
Keywords
Tuberculosis, Support vector machines, Medical diagnosis, Classification.
Introduction
Tuberculosis disease manifests itself if tuberculosis microbes get active and start to reproduce after entering the body by various ways. When left untreated, the disease causes damage at the affected organ and can lead to death.
Tuberculosis is present in all countries worldwide. Today, it is still the most fatal contagious disease caused by a single microbe. Annually 9.2 million people contract this disease, whereas 1.6 million die because of it. It comprises 2.5% of all diseases and 26% of preventable deaths in the world. Number of tuberculosis patients tends to increase worldwide (WHOWorld Health Organization). It is estimated that 12-15 million people in Turkey are infected, that is, they carry the tuberculosis microbe which has not caused disease yet. Approximately ten percent of these people will suffer from tuberculosis in some period of their lives.
After tuberculosis microbes enter the body, time to develop disease and chance of developing the disease vary among individuals. Those with lower body resistance and small children are most likely to develop the disease. The disease can develop soon after the microbe is contracted, or it can manifest itself after decades [1].
Symptoms of the disease generally start mild and progress slowly. For this reason many patients delay attending to a physician. Some patients attribute their coughing to smoking or another reason, and do not attend a physician. This condition can lead to wrong diagnosis and treatment.
Patient complaints and findings on chest X-ray raise suspicion for the disease. In suspected cases, definitive diagnosis of tuberculosis relies on demonstration of the bacillus in microscopy and culture in growth media. Sputum or rarely some other samples obtained from the patient are examined in the laboratory and the definite diagnosis is made [1-8].
There have been several studies recently to aid physicians in decision-support systems and diagnosis of the disease. In these studies, expert systems and different artificial intelligence techniques for classification systems in medical diagnosis is increasing gradually. As for other clinical diagnosis problems, artificial diagnostic systems have been used for tuberculosis diagnosis problem [1-8].
One of the several successful machine learning algorithms that have been developed to solve classification problems in the recent years is the Support Vector Machines. These algorithms have been successfully implemented in solving several classification problems, and have been recognized due to their high generalization performance.
The most important advantage of Support Vector Machines [9,10] is that they resolve classification problem by converting it to their own squared optimization problem. Thus, number of processes during learning period for solving the problem is reduced, and it takes less time to reach resolution compared to other algorithms [11]. It provides great advantage particularly for high-volume datasets. Furthermore, it is more successful than the other algorithms regarding classification performance, computing complexity and practicability, since it is optimization based [12].
In this study, we developed a system that diagnoses whether a patient has tuberculosis using database [1-6] established with SVM algorithm earlier. We compared obtained results to other studies which used same data with different algorithms for diagnosis of tuberculosis
Related Work
There are few studies related to automated diagnosis of tuberculosis using different artificial intelligence techniques. Most of these studies have high classification accuracy rates. These studies are mentioned below:
Asha et al. developed a hybrid model by combining k-means and other classification algorithms, namely SVM, C4.5 Decision Tree, Naive Bayes, K-NN, and Bagging Random Forest algorithms. They obtained the highest accuracy rate as 98.7% with SVM algorithm in their study [13].
Elveren and Yumusak used multilayer neural network (MLNN) with two hidden layers and a genetic algorithm for diagnosis of tuberculosis. They obtained 94.88% success rate in their study [14].
Dongardive et al. used the identification tree (IDT) for diagnosis of pulmonary tuberculosis and achieved 93% success rate in diagnostic accuracy. They used 250 patient data in their study in accordance with clinical data that they obtained from a local hospital in Mumbia [15].
Er et al. used multilayer neural network algorithm which is an artificial neural network model, for the diagnosis of tuberculosis using our same dataset in 2010. They used 150 patient data in their study and they achieved 94.88% success rate in diagnostic accuracy [4].
In their study in 2010, Er et al. used multilayer neural network algorithm which is an artificial neural network model, for the diagnosis of tuberculosis. They used 38 parameters present in patient discharge reports of 357 infected and normal patients that they obtained from a local hospital as input, and designed the output as either tuberculosis is present or absent. They achieved 95.8% success rate in diagnostic accuracy [5].
Ucar et al. used ANFIS (Adaptive-Network-based Fuzzy Interference System), which is a mixture of artificial neural network and fuzzy systems, with 503 different disease data for diagnosis of tuberculosis. They achieved 97% classification success with the help of ANFIS [16].
Theoretical backgrounds: Support Vector Machine (SVM)
In this section, we describe the SVM developed by Cortes and Vapnik in 1995. On the basis of statistical learning theory, SVM were proposed for classification and regression purposes [17-20]. They have proven good capabilities for the classification of complex and large datasets [21].
SVM is a statistical classification method originally designed for binary classification. Given a set T={(X,Y),......,(Xm,ym)} of m training data, where Xi ϵ Rn representing the with training data and yi ϵ {-11} the class label of, SVM provides the optimal hyperplane f(x)=wT x-b that separates two classes. When the training data are linearly separable, this hyperplane separates two classes with no training error, and maximizes minimum distance from the training data to the hyperplane. In order to maximize this minimum distance, we need to classify correctly the vectors xi of the training set into two different classes yi, using the smallest norm of coefficients w. The maximum hyperplane problem can be formulated as the following Quadratic Programming (QP) optimization problem [21]:
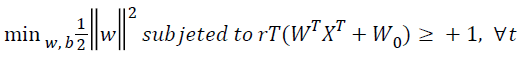 →(1)
→(1)
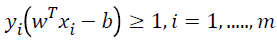 →(2)
→(2)
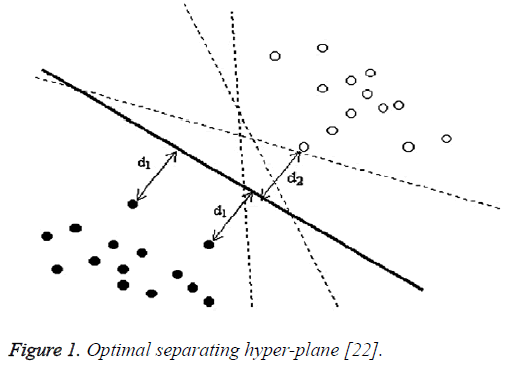
Figure 1 illustrates a linearly separable case, where data points of one category by black hole and data points of another category by empty hole are separated by the linear optimal separating hyper-plane (the solid straight line). There are actually an infinite number of hyper-planes that are able to partition the data points into two categories (as illustrated by the dashed lines on Figure 1). According to the SVM methodology, there will just be one optimal separating hyperplane. This optimal separating hyper-plane is lying half-way in between the maximal margin, where the margin is defined as the sum of distances of the hyper-plane to the support vectors. In the case as illustrated in Figure 1, the margin is d1+d2. The optimal separating hyper-plane is only determined by the closest data points of each category. These points are called Support Vectors (SVs). As only the SVs determine the optimal separating hyper-plane, there is a certain way to represent them for a given set of training points. It has been shown that the maximal margin can be found by minimizing as shown in Eq (1). Therefore, the optimal separating hyper-plane can be configured by minimizing Eq (1) under the constraint of Eq (2), that the training data points are correctly separated [22].
Figure 1: Optimal separating hyper-plane [22].
Methodology
Data description
We tested our system with the same data that has been used in similar studies previously. These data was obtained from chest diseases department of a hospital in Diyarbakir, which is a city located south of Turkey. The database that was used by the developed system comprises 50 patients diagnosed with tuberculosis and 100 disease-free individuals, totally accounting to 150 samples. System input was made by 38 properties that were present in patient discharge reports in each sample. These properties were as follows: ache on chest, weakness, complaint of cough, body temperature, dyspnoea on exertion, habit of cigarette, rattle in chest, pressure on chest, sputum, sound on respiratory tract, leucocyte, erythrocyte, thrombosis, haematocrit, haemoglobin, albumin 2, alkalis phosphatase 2 L, amylase, aspartate aminotransferase, alanine aminotransferase, bilirubin (total+direct), CK/creatinine kinase total, CK-MB, iron, gamma-glutamic transferase, glucose, HDL cholesterol, calcium, chlorine, blood urea nitrogen, cholesterol, potassium, sodium, creatinine, lactic dehydrogenase, total protein, uric acid, triglesid.
Diagnosis of the tuberculosis disease using SVM
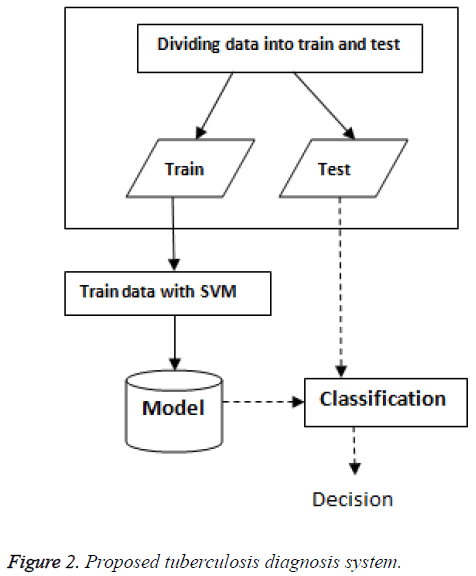
Figure 2 shows a summarizing of the proposed methodology. The objective of the tuberculosis diagnosis problem is to predict the property of a new patient (infected or not infected). In this study, SVM classifier with linear kernel was used for tuberculosis diagnosis.
In the beginning, the database is split into training-test partitions: The training data is used to create the model for training. After obtaining the predictive model, we conduct the prediction on each testing data.
Experimental results
The performance of our system is evaluated by using classification accuracy as statistical measures method. The classification accuracies obtained by this and best values of the referenced studies for tuberculosis disease same dataset were presented in Table 1.
| Study | Method (HL: hidden layer) |
Classification accuracy (%) |
|---|---|---|
| Same Tuberculosis Dataset [4] | GRNN (1-HL) | 93.18 |
| MLNN (BPwM, 1-HL) | 93.04 | |
| MLNN (LM, 1-HL) | 93.42 | |
| MLNN (BPwM, 2-HL) | 93.93 | |
| MLNN (LM,2-HL) | 95.08 | |
| Same Chest Dataset including Tuberculosis class[5] | MLNN (BPwM, 1-HL) | 84.00 |
| MLNN (BPwM, 2-HL) | 84.00 | |
| MLNN (LM, 1-HL) | 84.00 | |
| MLNN (LM, 2-HL) | 90.00 | |
| PNN | 88.00 | |
| LVQ | 84.00 | |
| GRNN | 86.00 | |
| RBF | 86.00 | |
| Same Tuberculosis Dataset [14] | MLNN with Genetic Algorithm | 94.88 |
| Our study Tuberculosis Dataset |
SVM | 96.68 |
Table 1: Average of classification accuracies of algorithms for tuberculosis disease diagnosis.
As shown in Table 1, in the study by Er et al., in which the same database was used with artificial neural network, highest success rate was achieved as 90% using MLNN with BP (Back-Propagation) algorithm in 2010 [4] and %95.08 using MLNN with LM (Levenberg-Marquardt) in 2010 [5]. In addition, they achieved success rates of 88%, 84%, 93.18% and 86% with PNN, LVQ, GRNN and RBF methods, respectively [4,5]. And in the study by Elveren & Yumusak, highest success rate was achieved as 94.88% using MLNN with our same dataset [14].
It is one of common performance measure in the literature defined as correct classified instances divided by the total number of instances with using different (input-output numbers) dataset [13].
As it is seen in the same table, we achieved 96.68% successful diagnostic accuracy rate using SVM method in this study. Furthermore, we saw that we achieved better and acceptable results than both MLNN and the other methods, namely PNN, LVQ, GRNN and RBF. These results can be explained by the effectiveness of SVM for tuberculosis diagnosis.
By way of conclusion, the following results should be mentioned:
In general, the classification accuracy obtained by this study is close to those obtained by reference studies [4-5,14]. The SVM not only present high accuracy rate but also reduces the training time. It is obtained that using SVM is a successful way to diagnose tuberculosis.
Conclusion
In this paper, we proposed the use of SVM for tuberculosis diagnosis. Experimental results are encouraging. It can be seen that SVM methods bring a significant performance to reach 96.68% with only a few minutes of runtime necessary for training. The results were also compared with the previous studies. The classification accuracy obtained by this study using SVM was better than those obtained in the previous studies. We can say that SVM could be successfully used for diagnosis of tuberculosis. In conclusion, practical implications of this study would be beneficial for physicians for the diagnosis of tuberculosis. This kind of systems could be directly used for prediction of the disease when used routinely in hospitals where there are no specialist doctors.
References
- Er O. Thoracic Disease Diagnosis using Flexible Computing and Bioinformatics Computing System. PhD Thesis, Sakarya University, 2009.
- Er O, Temurtas F. A Study on Chronic Obstructive Pulmonary Disease Diagnosis Using Multilayer Neural Networks. J Med Syst 2008; 32: 429.
- Er O, Sertkaya C, Temurtas F, Tanrikulu AC. A Comparative Study on Chronic Obstructive Pulmonary and Pneumonia Diseases Diagnosis using Neural Networks and Artificial Immune System. J Med Syst 2009; 33: 485.
- Er O, Temurtas F, Tanrikulu AC. Tuberculosis Disease Diagnosis Using Artificial Neural Networks. J Med Syst 2010; 34: 299.
- Er O, Yumusak N, Temurtas F. Chest diseases diagnosis using artificial neural networks. Exp Sys with Appl 2010; 37: 7648.
- Er O, Yumusak N, Temurtas F. Diagnosis of chest diseases using artificial immune system. Exp Sys with Appl 2012; 39: 1862.
- Er O, Tanrikulu AC, Abakay A. Use of artificial intelligence techniques for diagnosis of malignant pleural mesothelioma. Dic Med J 2015; 42: 5.
- Temurtas F. A comparative study on thyroid disease diagnosis using neural networks. Exp Sys with Appl 2009; 36: 944.
- Veropoulos K, Campbell C, Cristianini N. Controlling the sensitivity of support vector machines. In Proceed of the Int Join Conf on AI 1999; 55-60.
- Trafalis TB, Ince H. Support vector machine for regression and applications to financial forecasting. NeurNetwor 2000; 6: 6348.
- Osowski S, Siwek K, Markiewicz T. MLP and SVM networks–a comparative study. Proceed of the 6th Nor Sig ProcesSympos–NORSIG 2004; 9-11.
- Nitze I, Schulthess U,Asche H. Comparison of machine learning algorithms random forest, artificial neural network and support vector machine to maximum likelihood for supervised crop type classification. Proceed of the 4th GEOBIA, 2012; 35-40.
- Asha T, Natarajan S, Murthy KNB. A Data Mining Approach to the Diagnosis of Tuberculosis by Cascading Clustering and Classification. arXiv preprint arXiv 2011; 3.
- Elveren E, Yumusak N. Tuberculosis disease diagnosis using artificial neural network trained with genetic algorithm. J Medical Syst 2011; 35: 329.
- Dongardive J, Xavier A, Jain K, Abraham S. Classification and Rule-Based Approach to Diagnose Pulmonary Tuberculosis. AdvComput and Commun 2011; 328.
- Ucar T, Karahoca A, Karahoca D. Tuberculosis disease diagnosis by using adaptive neuro fuzzy inference system and rough sets. NeuComputApplicat 2013; 23:471.
- Meier TB, Desphande AS, Vergun S, Nair VA, Song J, Biswal BB, Meyerand ME, Birn RM, Prabhakaran V. Support vector machine classification and characterization of age-related reorganization of functional brain networks. Neuroimage 2012; 60: 601.
- Osowski, S, Linh TH, Tomasz M. Support vector machine-based expert system for reliable heartbeat recognition. Biomed Eng 2004; 51: 582.
- Daliri MR. Feature selection using binary particle swarm optimization and support vector machines for medical diagnosis. Biomed Eng-Biomed Tech 2012; 57: 395.
- Çomak E, Ahmet A, Ibrahim T. A decision support system based on support vector machines for diagnosis of the heart valve diseases. Comp in Biol and Med 2007; 37: 21.
- Cortes C, Vapnik V. Support-vector networks. Mach learn1995; 20: 273.
- Lee LH, Rajkumar R, Lo LH, Wan CH, Isa D. Oil and gas pipeline failure prediction system using long range ultrasonic transducers and Euclidean-Support Vector Machines classification approach. ExpSyst with Appl 2013; 40: 1925.