Research Article - Biomedical Research (2017) Volume 28, Issue 15
The methylated oligonucleotide-induced methylation of DKK3 promoter promotes the malignant properties of osteosarcoma cells
Shunguang Chen1,2, Shiqing Liu1* and Wanli Zhang1
1Department of Orthopedics, Renmin Hospital of Wuhan University, Hubei, PR China
2Department of Orthopedics, Jingzhou Central Hospital Affiliated to Tongji Medical College of Huazhong University of Science and Technology, Hubei, PR China
- *Corresponding Author:
- Shiqing Liu
Department of Orthopedics
Renmin Hospital of Wuhan University, PR China
Accepted on June 6, 2017
Abstract
Background: Deregulation of DKK3 has been reported in various cancers, and its abnormal methylation is closely associated with carcinogenesis. However, the methylation status and function of DKK3 in osteosarcoma have been poorly understood. This study investigated the effects of methylated oligonucleotide-induced methylation of DKK3 promoter on malignant properties of osteosarcoma cells.
Methods: SAOS-2 osteosarcoma cells were transfected with methylated or control oligonucleotides. Differences in the methylation status of the DKK3 promoter, proliferation, cell cycle, apoptosis, and metastasis of SAOS-2 cells were compared among the groups treated with different oligonucleotides.
Results: The Methylated Oligonucleotide (MON) successfully induced the methylation of the complementary CG sequences in the DKK3 promoter. Viable cells in the MON group were significantly more than that of the control groups. Consistently, in the MON group, the rate of S phase of cells was higher, while the rate of cellular apoptosis was lower than that in control groups. Moreover, the MON could also enhance the metastasis capability of SAOS-2 cells.
Conclusion: The methylated oligonucleotide induced the methylation of the DKK3 promoter, and exacerbated the proliferation and metastasis potential of SAOS-2 cells, suggesting that DKK3 promoter methylation could contribute to the malignant transformation of osteosarcoma. Taken together, these findings provide the evidence linking the methylation of the DKK3 promoter to osteosarcoma progression.
Keywords
DKK3, Methylation, Cell proliferation, Metastasis, Osteosarcoma.
Introduction
Osteosarcoma is a primary malignancy of bone that commonly happens in children and adolescents. It is highly aggressive, with the lung of the most common metastatic site [1]. Standardized application of neoadjuvant chemotherapy has increased the possibility of limb salvage, and contributed to the improved 5 y survival rate and life quality of osteosarcoma patients [2]. Despite intensive efforts have been devoted to osteosarcoma during the past decade, many patients still fail to achieve long-term disease free survival. The mechanisms underlying osteosarcoma progression currently are largely unknown. A better understanding of the underlying mechanisms is urgently required for improved treatment of osteosarcoma. Epigenetic modifications, including promoter methylation and chromatin condensation, have been proposed to contribute to osteosarcoma progression.
Dickkopf-related protein 3 (DKK3) is a 38 kDa secreted glycoprotein with an N-terminal signal peptide, which could not only elicit distinct intracellular roles, but also has secretary functions [3]. Aberrant Wnt signaling has been well established in the progression of many malignancies [4]. As one of the Wnt signaling pathway antagonists, DKK3 has been reported to be a potential tumor suppressor in various cancers, and its ectopic expression could suppress aggressive malignant properties, mostly resulting in reduced proliferation and metastasis through the reversion of Epithelial-Mesenchymal Transition (EMT) [5-8]. Lorsy has demonstrated that low DKK3 mRNA expression was significantly associated with reduced Recurrence Free Survival (RFS) of luminal and basal like breast cancer cases, and DKK3 re-expression in human breast cancer cell lines led to suppression of cell growth [9]. Transfection of DKK3 significantly reduced the invasion capacity and cell motility of SAOS-2 osteosarcoma cells, associated with cellular morphology changes consistent with a less invasive phenotype [8]. However, SAOS-2 cells with ectopical expression of Dkk-3 exhibited enhanced resistance to serum starvation and chemotherapy-induced cytotoxicity [8]. Recently, several studies have reported that DKK3 might exert oncogenic function [10,11]. The head and neck, and oral squamous cell carcinoma (HNSCC/OSCC) patients with DKK3 expression showed high metastasis rate and poorer survival, and DKK3 might contribute to the malignant progression in HNSCC/OSCC, through PI3K-Akt signaling other than the canonical Wnt signaling pathway [10,11]. To date, the function of DKK3 remains controversial and need to be clearly elucidated.
Recently, deregulation of DKK3 has been observed in multiple malignancies and influences the progression of carcinogenesis. DKK3 methylation is closely associated with the occurrence, progression, and prognosis of various cancers [12,13]. Xiang revealed that DKK3 methylation was detected in 78% of breast tumor samples, whereas DKK3 was rarely methylated in normal breast and surgical margin tissues, suggesting that DKK3 is methylated specifically in breast cancer [14]. In addition, the abnormal activity of Wnt/β-catenin signaling in hepatocellular carcinoma may be associated with the methylation of Dkk-3 [15]. Epigenetic downregulation of DKK3 also leads to docetaxel resistance in human Non-Small Cell Lung Cancer (NSCLC) [16]. However, the function and status of DKK3 methylation in carcinogenesis of osteosarcoma is rarely known.
Usually, DNA methyltransferases (DNMTs) could be used as a possible strategy to methylate the CpG islands of promoter. However, DNMT overexpression not only induces elevated methylation level of the target gene, but also increases total genome methylation level, which would cause extensive changes in gene expression. Recently, Yao et al. utilized a methylated oligonucleotide complementary to a region encompassing the human Insulin-Like Growth Factor 2 (IGF2) promoter to induce site-specific DNA methylation, which successfully induced hypermethylation of the promoter region and thus inhibition of gene transcription [17]. In addition, Ishii further confirmed the strategy that the methylated oligonucleotide induced methylation of Glutathione S-transferase P1-1 (GSTP1) promoter and suppressed its expression in lung adenocarcinoma cells [18].
Therefore, in this study, we aimed to induce methylation of DKK3 promoter by using the methylated oligonucleotide and then evaluated the effect of the methylation status of DKK3 on the malignant phenotype of SAOS-2 osteosarcoma cells. We found that induced methylation of DKK3 promoter exacerbated the proliferation and metastasis potential of SAOS-2 cells, suggesting that DKK3 promoter methylation could contribute to the malignant transformation of osteosarcoma. Taken together, these findings provide the evidence linking methylation of DKK3 promoter to tumor progression in human osteosarcoma.
Materials and Methods
Reagents and cells
SAOS-2 osteosarcoma cells were purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China). Fetal calf serum (FBS) and Dulbecco's Modified Eagle Medium (DMEM) were purchased from HyClone (Logan, UT, USA).
Effectene transfection reagents were purchased from QIAGEN China Co., Ltd. (Shanghai, China). Methylation detection kit was purchased from Beijing Tianmo Science and Technology Development Co., Ltd. (Beijing, China), and the CCK-8 kit was purchased from Beyotime Biotechnology (Haimen, China). Primers and methylated oligonucleotides were synthesized and purified by Takara Biotechnology Co., Ltd (Dalian, China), and sequencing was performed by Beijing Genomics Institute. SAOS-2 osteosarcoma cells were cultured in 90% DMEM supplemented with 10% FBS at 37°C in an incubator with an atmosphere of 5% CO2.
Oligonucleotide sequences
Sequence of the DKK3 promoter was obtained from GenBank (http://www.ncbi.nlm.nih.gov/gene) and DBTSS (http://dbtss.hgc.jp/) databases. Methylated and non-methylated oligonucleotides were designed based on those reported in previous studies [17-20]; sequences of these oligonucleotides are listed in Table 1. The map of the CpG islands of the DKK3 promoter targeted by the specific methylated oligonucleotide is shown in Supplementary Figure 1. Oligonucleotides in the Methylated Oligonucleotide (MON) group were complementary to the DKK3 promoter and contained methylated CG (mCG). Oligonucleotides in UMON group were complementary to the DKK3 promoter but did not contain mCG. Oligonucleotides in CON1 group had a random sequence, did not contain mCG, and were not complementary to the DKK3 promoter. Oligonucleotides in CON2 group contained mCG but were not complementary to the DKK3 promoter. CON3 group was a control group that did not contain oligonucleotides.
| Groups | Base sequence of methylated oligonucleotides |
|---|---|
| MON group | GGmCGGGGmCGCTmCGAGTAGGACCmCGAmCGCC |
| UMON group | GGCGGGGCGCTCGAGTAGGACCCGACGCC |
| CON1 group | TTACGCTTAAACGCTTGCGTAGAACGCTA |
| CON2 group | TTAmCGCTTAAAmCGCTTGmCGTAGAAmCGCTA |
| CON3 group | - |
Table 1. The sequences of the oligonucleotides in the MON, UMON, CON1, CON2, and CON3 groups.
Transfection of oligonucleotides
SAOS-2 osteosarcoma cells were subcultured, and transfection was performed when the cells reached approximately 50% confluency. Cells in the MON, UMON, CON1, and CON2 groups were transfected with the respective oligonucleotides and the CON3 group was only treated with the transfection reagent without any oligonucleotides.
Detection of the methylation status of the DKK3 promoter
Genomic DNA of cells in the MON, UMON, CON1, CON2, and CON3 groups was extracted. Methylation status of the DKK3 promoter was determined by performing bisulfite sequencing PCR. Methylation primers (forward primer, AGTTTAGTTTTTTTTGGTGGATGTG; reverse primer, AAACCCCAACTCACCTAAACTCTAT) were designed using MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) [21]. PCR conditions were as follows: initial denaturation at 95°C for 10 min; 35 cycles of denaturation at 95°C for 1 min, annealing at 63°C for 30 s, and extension at 72°C for 30 s; and final extension at 72°C for 10 min. PCR products were cloned by performing TA cloning, and their sequences were analysed using BiQ Analyzer.
The cell viability assay
The cell proliferation was detected using the CCK-8 assay according to the producer's protocol (Beyotime Biotechnology, Haimen, China). Briefly, cells were seeded in 96-well plates with a density of 5 × 103 cells per well overnight. The cells were treated with different oligonucleotides, and after different periods (0~7 d), the cell proliferation was evaluated by adding CCK-8 solution to each well of the plate. After incubation for 1 h, the absorbance was measured at 450 nm using a Multiskan MK3Microplate Reader (Thermo Scientific, USA). The absorbance was obtained by subtracting the absorbance of the blank control group from that of the experimental groups. The proliferation curves of d 0 to 7 (D0-D7) were plotted using absorbance as the ordinate and time as the abscissa.
Detection of cell cycle and apoptosis
Flow cytometry was used to identify cell cycle distribution and cell apoptosis. Briefly, SAOS-2 osteosarcoma cells were seeded in a six-well plate and transfected with different oligonucleotides after overnight culture. Cells were harvested by trypsinization and suspended in PBS containing 0.2 mg/ml propidium iodide, 1 mg/ml RNase, and 0.1% Triton X-100 for cell cycle. After 30 min incubation, the cell suspension was analysed with a FACS Caliber flow cytometer using CellQuest software (Becton Dickinson Co. Chino Hills, CA). The Proliferation Index (PI) was calculated using the following formula: PI=(S+G2/M)/(G0/G1+S+G2/M) × 100%. To quantify cellular apoptosis, transfected SAOS-2 osteosarcoma cells were stained with an annexin V staining Kit (Bestbio, Beijing, China) according to the manufacturer’s protocol and analysed by flow cytometry.
In vitro migration and invasion assay
In vitro migration and invasion assays were performed by placing cells into the upper chamber of a transwell (BD Bioscience) without or with Matrigel, under serum-free conditions. Medium supplemented with 10% FBS was used as a chemo attractant in the lower chamber. Briefly, SAOS-2 osteosarcoma cells were seeded in a six-well plate, and transfected with different oligonucleotides after overnight culture. Twenty-four hours after transfection, the cells were transferred to the transwell. After incubation for 48 h, cells remaining on the upper chamber were removed with a cotton swab, while cells adhering to the lower membrane were stained with 0.1% crystal violet and photographed with an inverted microscope.
Statistical analysis
Statistical analyses were conducted using SigmaPlot 12.0. One-way ANOVA with the Dunnett’s post-test was used to compare the means of three or more groups. P<0.05 was considered statistically significant at an inspection level (α) of 0.05.
Results
The methylated oligonucleotides induce the methylation of DKK3 promoter
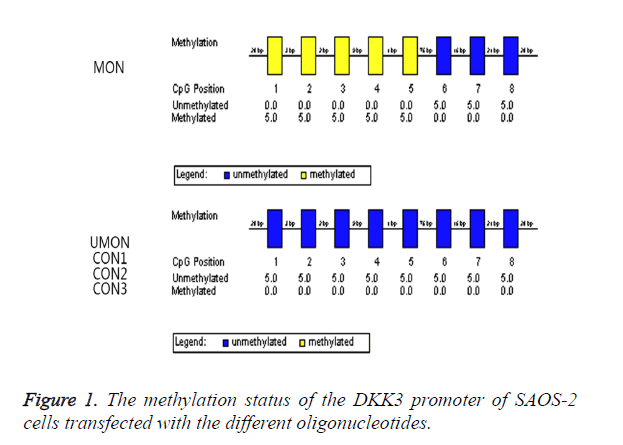
We attempted to induce the methylation of the CpG island of the DKK3 promoter by using MON. As shown in Figure 1, the oligonucleotides were complementary to a part of this promoter region including CpGs which were numbered as 1-5. The methylation status in the promoter region was analysed by a bisulfite-sequencing method. The CpGs numbered as 1-5 became methylated in 100% of clones from the cells transfected with MON, whereas the methylation status of the CpGs numbered as 6-8 was not changed by MON. In contrast, the CpGs numbered as 1-8 remained to be unmethylated in the UMON, CON1, CON2, and CON3 control groups. This indicated that the CpG island numbered as 1-8 of the DKK3 promoter in SAOS-2 was not methylated, and the MON successfully induced the methylation of the complementary CG sequences in the promoter but oligonucleotides in the control groups did not.
The methylated oligonucleotides promote the proliferation of SAOS-2 cells
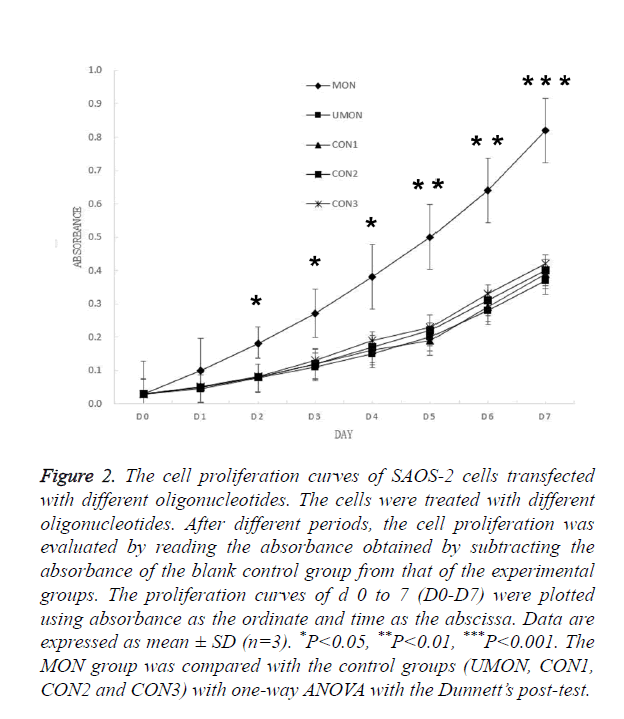
Having demonstrated that MON successfully induced the methylation of the DKK3 promoter, we examined whether MON could alter survival capacity of SAOS-2 osteosarcoma cells. As shown in Figure 2, no significant differences were observed in the D0–D7 absorbance of cells among the control groups (UMON, CON1, CON2, and CON3). Though the D0- D1 absorbance of cells in the MON group was not significantly different from that of cells in the control groups (P>0.05), the D2-D7 absorbance of cells in the MON group were significantly higher than the control groups (P<0.05). The result indicated that the MON, which induced the methylation of the DKK3 promoter, could promote the proliferation of SAOS-2 cells.
Figure 2. The cell proliferation curves of SAOS-2 cells transfected with different oligonucleotides. The cells were treated with different oligonucleotides. After different periods, the cell proliferation was evaluated by reading the absorbance obtained by subtracting the absorbance of the blank control group from that of the experimental groups. The proliferation curves of d 0 to 7 (D0-D7) were plotted using absorbance as the ordinate and time as the abscissa. Data are expressed as mean ± SD (n=3). *P<0.05, **P<0.01, ***P<0.001. The MON group was compared with the control groups (UMON, CON1, CON2 and CON3) with one-way ANOVA with the Dunnett’s post-test.
The methylated oligonucleotides increase the rate of S phase and decrease apoptosis in SAOS-2 cells
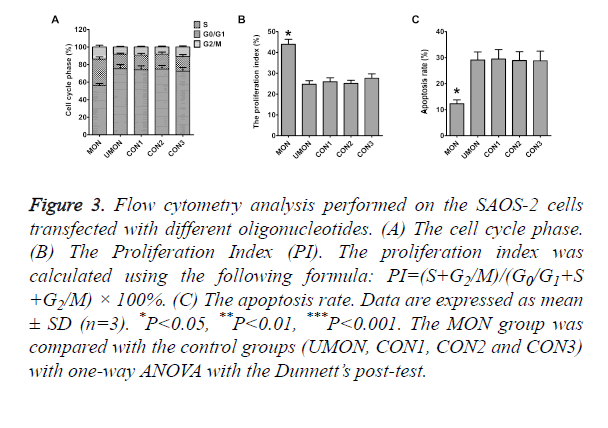
To reveal the mechanisms through which the MON promoted the proliferation of SAOS-2 cells, flow cytometry was performed to quantitative the population of apoptotic cells and cell cycle distribution. As shown in Figure 3, differences in the rates of G0/G1 phase, S phase, G2/M phase, Proliferation Index (PI), and apoptosis were not statistically significant among the cells in the control groups (P>0.05). The rates of S phase, G2/M phase, and PI of cells in the MON group were significantly higher than those in the control groups (P<0.05) (Figures 3A and 3B). The MON also strikingly decreased the apoptosis of SAOS-2 cells compared with the control groups (P<0.05) (Figure 3C).
Figure 3. Flow cytometry analysis performed on the SAOS-2 cells transfected with different oligonucleotides. (A) The cell cycle phase. (B) The Proliferation Index (PI). The proliferation index was calculated using the following formula: PI=(S+G2/M)/(G0/G1+S +G2/M) × 100%. (C) The apoptosis rate. Data are expressed as mean ± SD (n=3). *P<0.05, **P<0.01, ***P<0.001. The MON group was compared with the control groups (UMON, CON1, CON2 and CON3) with one-way ANOVA with the Dunnett’s post-test.
Methylated oligonucleotides exacerbate the metastatic potential of SAOS-2 cells
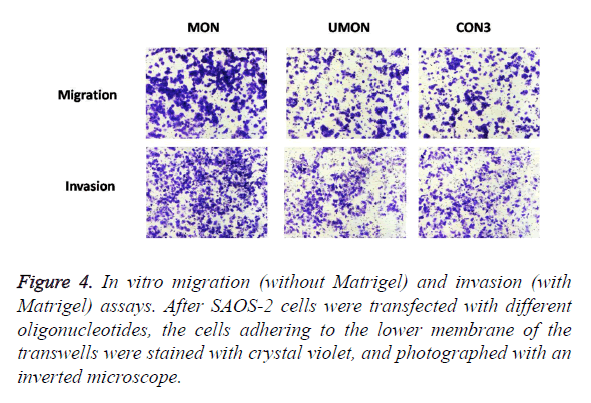
To evaluate the effect of the MON on the metastatic capability of SAOS-2 cells, oligonucleotides were transfected into SAOS-2 cells, and migration and invasion assay in vitro were performed (Figure 4). MON treatment could significantly increase the migration and invasion of SAOS-2 cells, compared with the control UMON. However, the UMON did not influence the metastatic potential of SAOS-2 cells compared with CON3. The result indicated that the MON could promote the metastasis of SAOS-2 cells.
Discussion
Epigenetics involves the study of methylation and acetylation, and mechanisms regulating these processes do not rely on changes in the base sequence of genes. Methylation of gene promoters is the most common and important regulatory mechanism of epigenetics [22]. Methylation or binding of proteins to methylated CpG islands inhibits the binding of transcription factors to gene promoters, decreases gene expression, and regulates gene expression before transcription [23]. Increasing genes are found with hypermethylation inhibition in tumors [24]. Hypermethylation was detected in tumor tissues such as gastrointestinal tumors, respiratory tract neoplasms [25,26]. Abnormal demethylation is closely associated with the development of tumors and is regarded as a hallmark for evaluating prognosis [27,28]. Deregulation of DKK3 has been reported in multiple malignancies, and its abnormal methylation is closely associated with the progression of carcinogenesis [29-34]. However, the methylation status and function of DKK3 in osteosarcoma has not been clarified. Our present study showed that induced methylation of DKK3 promoter promoted the proliferation of SAOS-2 osteosarcoma cells, as reflected by increased rate of S phase and decreased apoptosis, and exacerbated migration and invasion capability. The results indicated that DKK3 promoter methylation could contribute to the malignant transformation of osteosarcoma, and promote the development and progression of osteosarcoma. In the future study, we should further compare the methylation status of the DKK3 promoter in patients of osteosarcoma with healthy people.
Methylated oligonucleotides are artificially synthesized to contain sequences complementary to the target gene. Hemimethylated DNA is formed once methylated oligonucleotides anneal with the target gene and methylate complementary sequences in the gene in the presence of DNA methyltransferase. Subsequently, the methylated DNA strand and its complementary strand undergo replication, which eventually decreases the expression of the target gene and causes the gene to lose its normal function [35]. This strategy could specifically induce the promoter methylation of the target gene, and avoid causing extensive changes in gene expression. Recently, methylated oligonucleotides were successfully used to inactivate genes such as GSTP1 and IGF2 [17,18]. In the present study, we designed the following oligonucleotides based on the criteria for designing the methylated and control oligonucleotides. In the MON group, treatment of cells with the methylated oligonucleotides resulted in the methylation of target CG sites in the DKK3 promoter. In contrast, no methylation of the DKK3 promoter was observed in cells in the control groups. These results indicated that the DKK3 promoter in SAOS-2 osteosarcoma cells was not methylated at the CG sites and that the complementary methylated oligonucleotides successfully induced the methylation of these sites while control oligonucleotides did not.
In contrast to the traditional members of the DKK family, DKK3 does not only modulate the Wnt/β-catenin pathway, but also has distinct intracellular signaling partners, which are also closely associated with tumorigenesis [7]. A study showed that DKK3 promoted pathogenic stromal remodeling in benign prostatic hyperplasia and prostate cancer via enhancing PI3K/AKT signaling [36]. In the present study, the viable cells in the MON group was significantly higher than that of cells in the control groups, accompanied with increased rate of S phase and decreased apoptosis. Moreover, the MON could also enhance the metastasis capability of osteosarcoma cells. The results indicated that induced methylation of the DKK3 promoter promoted the malignant phenotype, maybe dependent or independent on the modulation of Wnt/β-catenin pathway. Thus, this study simulated the abnormal methylation of the DKK3 promoter occurring in tumor cells and highlighted the contribution of the effect of DKK3 methylation to the mechanisms underlying osteosarcoma progression.
Acknowledgements
This work was supported by the 863 program projects (project number: 2012AA020809).
References
- Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol 2015; 33: 3029-3035.
- Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am 2016; 47: 283-292.
- Lee EJ, Jo M, Rho SB, Park K, Yoo YN, Park J, Chae M, Zhang W, Lee JH. Dkk3, downregulated in cervical cancer, functions as a negative regulator of beta-catenin. Int J Cancer 2009; 124: 287-297.
- Valencia A, Roman-Gomez J, Cervera J, Such E, Barragán E, Bolufer P, Moscardó F, Sanz GF, Sanz MA. Wnt signaling pathway is epigenetically regulated by methylation of Wnt antagonists in acute myeloid leukemia. Leukemia 2009; 23: 1658-1666.
- Ochiai K, Watanabe M, Azakami D, Michishita M, Yoshikawa Y, Udagawa C, Metheenukul P, Chahomchuen T, Aoki H, Kumon H, Morimatsu M, Omi T. Molecular cloning and tumour suppressor function analysis of canineREIC/Dkk-3 in mammary gland tumours. Vet J 2013; 197: 769-775.
- Zenzmaier C, Heitz M, Klocker H, Buck M, Gardiner RA, Berger P. Elevated levels of Dickkopf-related protein 3 in seminal plasma of prostate cancer patients. J Transl Med 2011; 9: 193.
- Veeck J, Dahl E. Targeting the Wnt pathway in cancer: the emerging role of Dickkopf-3. Biochim Biophys Acta 2012; 1825: 18-28.
- Hoang BH, Kubo T, Healey JH, Yang R, Nathan SS, Kolb EA, Mazza B, Meyers PA, Gorlick R. Dickkopf 3 inhibits invasion and motility of SAOS-2 osteosarcoma cells by modulating the Wnt-beta-catenin pathway. Cancer Res 2004; 64: 2734-2739.
- Lorsy E, Topuz AS, Geisler C, Stahl S, Garczyk S. Loss of Dickkopf 3 Promotes the Tumorigenesis of Basal Breast Cancer. PLoS One 2016; 11: 0160077.
- Katase N, Nishimatsu SI, Yamauchi A, Yamamura M, Terada K, Itadani M, Okada N, Hassan NMM, Nagatsuka H, Ikeda T, Nohno T, Fujita S. DKK3 overexpression increases malignant properties of head and neck squamous cell carcinoma cells. Oncol Res 2017.
- Zhou S, Zhu Y, Mashrah M, Zhang X, He Z, Yao Z, Zhang C, Guo F, Hu Y, Zhang C. Expression pattern of DKK3, dickkopf WNT signaling pathway inhibitor 3, in the malignant progression of oral submucous fibrosis. Oncol Rep 2017; 37: 979-985.
- Katase N, Lefeuvre M, Tsujigiwa H, Fujii M, Ito S, Tamamura R, Buery RR, Gunduz M, Nagatsuka H. Knockdown of Dkk-3 decreases cancer cell migration and invasion independently of the Wnt pathways in oral squamous cell carcinoma-derived cells. Oncol Rep 2013; 29: 1349-1355.
- Katase N, Lefeuvre M, Gunduz M, Gunduz E, Beder LB, Grenman R, Fujii M, Tamamura R, Tsujigiwa H, Nagatsuka H. Absence of Dickkopf (Dkk)-3 protein expression is correlated with longer disease-free survival and lower incidence of metastasis in head and neck squamous cell carcinoma. Oncol Lett 2012; 3: 273-280.
- Xiang T, Li L, Yin X, Zhong L, Peng W, Qiu Z, Ren G, Tao Q. Epigenetic silencing of the WNT antagonist Dickkopf 3 disrupts normal Wnt/β-catenin signalling and apoptosis regulation in breast cancer cells. J Cell Mol Med 2013; 17: 1236-1246.
- Liang L, He H, Lv R, Zhang M, Huang H, An Z, Li S. Preliminary mechanism on the methylation modification of Dkk-1 and Dkk-3 in hepatocellular carcinoma. Tumour Biol 2015; 36: 1245-1250.
- Tao L, Huang G, Chen Y, Chen L. DNA methylation of DKK3 modulates docetaxel chemoresistance in human nonsmall cell lung cancer cell. Cancer Biother Radiopharm 2015; 30: 100-106.
- Yao X, Hu JF, Daniels M, Shiran H, Zhou X, Yan H, Lu H, Zeng Z, Wang Q, Li T, Hoffman AR. A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma. J Clin Invest 2003; 111: 265-273.
- Ishii T, Fujishiro M, Masuda M, Teramoto S, Matsuse T. A methylated oligonucleotide induced methylation of GSTP1 promoter and suppressed its expression in A549 lung adenocarcinoma cells. Cancer Lett 2004; 212: 211-223.
- Li XQ, Pei DS, Qian GW, Yin XX, Cheng Q, Li LT, Li HZ, Zheng JN. The effect of methylated oligonucleotide targeting Ki-67 gene in human 786-0 renal carcinoma cells. Tumour Biol 2011; 32: 863-872.
- Li HL, Ma AN. Induction of apoptosis of non-small cell lung cancer by a methylated oligonucleotide targeting survivin gene. Cancer Gene Ther 2010; 17: 441-446.
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18: 1427-1431.
- Seven D, Yavuz E, Kilic E, Baltaci E, Karaman E. DLEC1 is not silenced solely by promoter methylation in head and neck squamous cell carcinoma. Gene 2015; 563: 83-86.
- Kon T, Yoshikawa N. Induction and maintenance of DNA methylation in plant promoter sequences by apple latent spherical virus-induced transcriptional gene silencing. Front Microbiol 2014; 5: 595.
- Lattanzio L, Borgognone M, Mocellini C, Giordano F, Favata E. MGMT promoter methylation and glioblastoma: a comparison of analytical methods and of tumor specimens. Int J Biol Markers 2015; 30: 208-216.
- Dai Z, Jin Y. Promoter methylation of the DLCâ€1 gene and its inhibitory effect on human colon cancer. Oncol Rep 2013; 30: 1511-1517.
- Huang YZ, Wu W, Wu K, Xu XN, Tang WR. Association of RASSF1A promoter methylation with lung cancer risk: a meta-analysis. Asian Pac J Cancer Prev 2014; 15: 10325-10328.
- Dong X, Liu RY, Chen WD. Correlation of promoter methylation in the MGMT gene with glioma risk and prognosis: a meta-analysis. Mol Neurobiol 2015; 52: 1887.
- Wu L, Wang F, Xu R, Zhang S, Peng X. Promoter methylation of BRCA1 in the prognosis of breast cancer: a meta-analysis. Breast Cancer Res Treat 2013; 142: 619-627.
- Kobayashi K, Ouchida M, Tsuji T, Hanafusa H, Miyazaki M, Namba M, Shimizu N, Shimizu K. Reduced expression of the REIC/Dkk-3 gene by promoter- hypermethylation in human tumor cells. Gene 2002; 282: 151-158.
- Yu J, Tao Q, Cheng YY, Lee KY, Ng SS, Cheung KF, Tian L, Rha SY, Neumann U, Rocken C, Ebert MP, Chan FK, Sung JJ. Promoter methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is associated with poor survival in gastric cancer. Cancer 2009; 115: 49-60.
- Veeck J, Bektas N, Hartmann A, Kristiansen G, Heindrichs U, Knuchel R, Dahl E. Wntsignalling in human breast cancer: expression of the putative Wnt inhibitor Dickkopf-3 (DKK3) is frequently suppressed by promoter hypermethylation in mammary tumours. Breast Cancer Res 2008; 10: R82.
- Yang B, Du Z, Gao YT, Lou C, Zhang SG, Bai T, Wang YJ, Song WQ. Methylation of Dickkopf-3 as a prognostic factor in cirrhosis-related hepatocellular carcinoma. World J Gastroenterol 2010; 16: 755-763.
- Lodygin D, Epanchintsev A, Menssen A, Diebold J, Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res 2005; 65: 4218-4227.
- Chim CS, Pang R, Fung TK, Choi CL, Liang R. Epigenetic dysregulation of Wnt signaling pathway in multiple myeloma. Leukemia 2007; 21: 2527-2536.
- Place TL, Fitzgerald MP, Venkataraman S, Vorrink SU, Case AJ. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PLoS One 2011; 6: 14617.
- Zenzmaier C, Sampson N, Plas E, Berger P. Dickkopf-related protein 3 promotes pathogenic stromal remodeling in benign prostatic hyperplasia and prostate cancer. Prostate 2013; 73: 1441-1452.