- Journal of RNA and Genomics (2007) Volume 3, Issue 1
The generation of small RNAs; who needs Dicer?
| James S Parker* Department of Biochemistry, South Parks Road, University of Oxford, Oxford OX1 3QU, UK, |
| Email:james.parker@bioch.ox.ac.uk, Tel: +44 (0)1865 275372, Fax: +44 (0)1865 275259 |
| (Received 26 May 2007; Published online 27 May 2007) |
Visit for more related articles at Journal of RNA and Genomics
Abstract
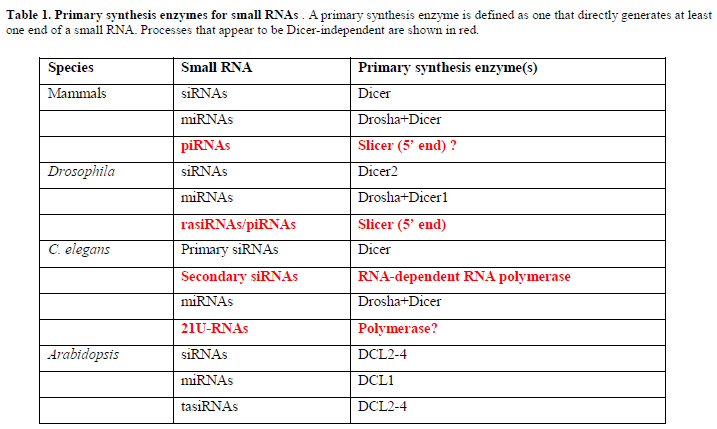
| Up until 2006 it was believed that most, if not all, small RNAs in RNA silencing were synthesized via members of the Dicer-like RNase III endonuclease family. In animals, small interfering RNAs (siRNAs) are cleaved from long double-stranded RNA by Dicer enzymes, and microRNAs (miRNAs) are released from genome-encoded stem-loop precursors through the sequential action of Droshas and Dicers. In plants, members of the Dicer-like (DCL) family function together in the biosynthesis of siRNAs, miRNAs and trans-acting siRNAs (tasiRNAs). Now, large-scale small RNA sequencing projects and improved methods for the characterisation of small RNAs have revealed two, perhaps three, new and distinct Dicer-independent mechanisms for the generation of small RNAs. |
| piRNAs are a new class of small RNAs found in the germ cells of higher animals (mammals, zebrafish, Drosophila – here termed rasiRNAs), one of whose key roles appears to be the suppression of parasitic mobile genetic elements, or transposons. (It is unclear why this regulation is required specifically in germ cells: whether the germ cell nuclear environment enhances the activity of transposons, or whether germ cells are particularly vulnerable to transposons.) piRNAs associate specifically with the Piwisubfamily of Argonaute proteins, including Mili and Miwi in mammals, and Piwi, Aubergine and Ago3 in Drosophila. |
| Recently, analysis of the piRNAs associated with the three Drosophila Piwi-subfamily proteins has led to some remarkable conclusions as to the mode of generation of piRNAs (Brennecke et al, 2007; Gunawardane et al, 2007). These studies find two distinct populations of piRNAs: 1) Those associated with Piwi or Aubergine, which display a preference for uracil at position 1 (as measured from the 5’ end) and are derived largely from strands antisense to transposon sequences, and 2) Those associated with Ago3, which display a preference for adenine at position 10 and are derived largely from sense strands (i.e., transposon transcripts). This is the relationship that would be expected if the Piwi and Aubergine associated RNAs, via the slicing activity of Piwi and Aubergine, were responsible for the generation of the 5’ ends of the Ago3-associated RNAs, and vice-versa. Furthermore, pairs of piRNAs are found that share complementarity over their first ten nucleotides, consistent with this linked relationship. |
| So, a model emerges whereby the 5’ end of a piRNA is determined via the slicer activity of Argonaute, not the endonuclease activity of Dicer or Drosha. This scheme in the germ cells of Drosophila results in the consumption of transposon transcripts, thereby leading to the suppression of transposon activity. Clearly also, the model is an amplification loop which responds on demand to nascent transposon activity. Unresolved questions remain however, most notably the identification of the mechanism that specifies the 3’ ends of piRNAs. The model also requires initiator piRNAs, although there is some suggestion that piRNA-Aubergine complexes may be deposited in the developing germline during oogenesis. |
| The RNAi response in C. elegans is unusual in animals is so far as it appears to be a two-phase response. The first phase corresponds to the production of primary siRNAs, which are derived from Dicer cleavage of a doublestranded RNA silencing trigger. The second phase, which is characterised by amplification of the silencing signal and spreading away from the initial site of silencing (transitive RNAi), is mediated by a distinct class of small RNAs, known as secondary siRNAs. The mode of biosynthesis of secondary siRNAs has until recently been unclear, although known to be dependent on RNA-dependent RNA polymerase (RdRP) activity. |
| Two recent papers from the Fire and Plasterk laboratories address the nature of C. elegans secondary siRNAs (Pak and Fire, 2007; Sijen et al, 2007). A remarkable finding is that these RNAs, unlike all other small RNAs in RNA silencing so far analysed, possess a 5’ tri-phosphate group. Previously, a defining feature of siRNAs and miRNAs had been a 5’ mono-phosphate group, produced as a result of Dicer or Drosha cleavage and recognised by specific binding pocket in the PIWI domain of Argonaute. Together with the finding that secondary siRNAs appear to be derived almost exclusively from the antisense strand, the results lead both groups to the proposal that each secondary siRNA in C. elegans is synthesized individually by RdRP. This differs markedly from the mechanism of secondary siRNA generation in plants, where RdRP generates a long complementary strand to form double-stranded RNA, which is then cleaved by Dicer. |
| Intriguingly, secondary siRNAs in C. elegans associate with members of a distinct and less well characterised subfamily of Argonaute proteins (Yigit et al, 2006). The novel biogenesis mechanism of secondary siRNAs may be reflected in the structures of these proteins. First, this subfamily have an atypical 5’ binding motif, whereby a tyrosine residue, that in other subfamilies contacts the 5’ monophosphate group and the base of the 5’ nucleotide, is replaced by histidine. Second, the sub-family is also unusual in that almost all members lack the catalytic slicer DDH residues. This may be a reflection of the fact that secondary siRNAs are created as single-stranded RNAs, so that slicing is not required for cleavage and release of a complementary passenger strand. Of course, function of the Argonaute-secondary siRNA complex will not involve slicing. |
| A third distinct biosynthesis mechanism may operate for a novel class of small RNAs found in C. elegans, termed 21U-RNAs (Ruby et al 2006). These RNAs, identified through high-throughput sequencing of small RNAs from C. elegans, are precisely 21 nucleotides long with a uridine 5’ monophosphate and a modified 3’ terminal ribose These RNAs appear to be encoded in clusters in the genome, but intriguingly each coding sequence is associated with a conserved upstream sequence motif. The authors suggest each 21U-RNA may be the product of an individual RNA polymerase transcription event. |
| The new diversity of mechanisms for the generation of small RNAs may come as a surprise to those involved in RNA silencing. It raises issues regarding the assembly of the guide RNA-Argonaute complex. Previously, it could be envisaged that small RNAs are incorporated into Argonaute via some common mechanism involving Dicer and other associated factors. Now we may contemplate three possibilities: 1) several different mechanisms exist, 2) a common mechanism exists – involving Dicer – which incorporates a degree of plasticity, or 3) a common mechanism exists, not involving Dicer, about which we currently have little understanding. |
| Short Copy right Statement |
| This is an open access article, published under the terms of the Licence for Users available at http://www.libpubmedia.co.uk/ RNAiJ/LicenceForUsers.pdf. This licence permits noncommercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details. |
 |
References
|