Review Article - Asian Journal of Biomedical and Pharmaceutical Sciences (2022) Volume 12, Issue 89
TET proteins and their role in regulation of DNA methylation.
Teka Obsa Feyisa1*, Mezgebu Legesse1, Getahun Chala2, Tefera Belsty3, Mohammed Yusuf3 and Webshet Nebyu3
1Department of Medical Biochemistry, College of Health and Medical Sciences, Haramaya University, Harar, Ethiopia
2Department of Medical Physiology, college of Health and Medical sciences, Haramaya University, Harar, Ethiopia
3Department of Human Anatomy, College of Health and Medical science, Haramaya University, Harar, Ethiopia
- Corresponding Author:
- Teka Obsa Feyisa
Department of Medical Biochemistry
College of Health and Medical Sciences
Haramaya University, Harar
Ethiopia
E-mail: takobsi2008@ gmail.com
Received: 21-April-2022, Manuscript No. AABPS-22-3386; Editor assigned: 25-April-2022, PreQC No. AABPS-22-3386(PQ); Reviewed: 09-May-2022, QC No. AABPS-22-3386; Revised: 12-May-2022, Manuscript No. AABPS-22-3386(R); Published: 19-May-2022, DOI:10.35841/2249-622X.89.121
Citation: Teka OF, Mezgebu L, Getahun C, et al. TET proteins and their role in regulation of DNA methylation. Asian J Biomed Pharmaceut Sci. 2022;12(89):121
Abstract
Ten-Eleven Translocation (TET) proteins are enzymes that are capable oxidizing of 5-methylcytosine (5mC) into three biologically essential molecules: 5-carboxylcytosine (5caC), 5-formyl cytosine (5fC), and 5-hydroxymethyl cytosine (5hmC). In mammalian genes, TET proteins are categorized into three main members (TET1, TET2 and TET3), which basically contain a C-terminal catalytically active domain and a central region enriched with cysteine domain. Before the finding of TET proteins, DNA methylation was supposed to be irreversible epigenetic change. The discovery of the TET protein later revealed the DNA methylation process to be a reversible covalent modification catalyzed by DNA methyl transferase enzymes. In addition to regulating DNA methylation pathways, TET proteins play significant roles in controlling gene expression, epigenetic remodeling in stem cell segregation, embryogenesis, growth, and cancer. Beside the gene expression, the activities TET proteins can be altered by an array of cellular processes such as interaction with chromatin protein and other different small molecules. Here, we reviewed the current concepts regarding the structure of TET proteins, their role in DNA methylation, gene regulation, and their mutation which leads to development of certain cancer.
Keywords
DNA Methylation, Epigenetic modification, TET proteins, 5-methylcytosine.
Introduction
Ten-eleven translocation (TET) proteins that are capable oxidizing of oxidizing 5-methylcytosine (5mC) into three biologically essential molecules: 5- hydroxyl methyl cytosine (5hmC), 5-formyl cytosine (5fC) and 5-carboxylcytosine (5caC) [1-3]. The TET proteins were originally discovered and named as a result of the unusual ten–eleven translocation (t(q22; q23)) detected in acute myeloid and lymphocytic leukemia cases where by the mixed-lineage leukemia 1 (MLL1) gene located on chromosome 10 is fused with the TET1 gene positioned on chromosome 11 in humans [4-7]. Three TET genes (TET1, TET2 and TET3) that have been recognized in mammals encode for proteins capable of catalyzing the oxidation of 5mC in the DNA demethylation pathways [8].
DNA methylation is defined as the covalent modification of the nitrogenous base cytosine by addition of a methyl group (-CH3) to the fifth position of 5-CpG-3 dinucleotide linkage. This covalent modification of DNA is principally recognized at CpG rich region in the mammalian genomes. The addition of –CH3 to non-CpG dinucleotide is atypical, except for stem cells and mature neuron cells [5,9,10]. DNA methylation is one of the well-studied epigenetic events catalyzed by DNA methyl transferase (DNMTs) and has crucial role in normal development, [4,5,11,12]. Several scientific studies were conducted on the TET proteins and their role in the regulation of DNA methylation, but a comprehensive review is limited. Thus, this review was aimed at presenting critically organized and comprehensive concepts on the role of TET proteins in the regulation of DNA methylation.
The structure of TET Protein Members
As it has been mentioned by different studies mammalian genes contain three main members of TET proteins: TET1 located on 10q21.3, TET2 located on 4q24 and TET3 located on 2p13.1 of human chromosome. The results of many studies showed catalytically active C-terminal domains, cysteine rich regions, are found in all TET proteins, as well as a doublestranded β-helix (DSBH) fold distinctive of the ferrous iron (Fe (II)) and 2-oxoglutarate dependent deoxygenate super family [8,13]. These enzymes utilize Fe (II) that serves as a cofactor and 2- oxoglutarate as a co substrate for the enhancement their catalytic activities. The three TET genes identified in mammals have been proved to encode for proteins that are able to oxidize 5mC as a part of DNA de methylation pathway [14,15].
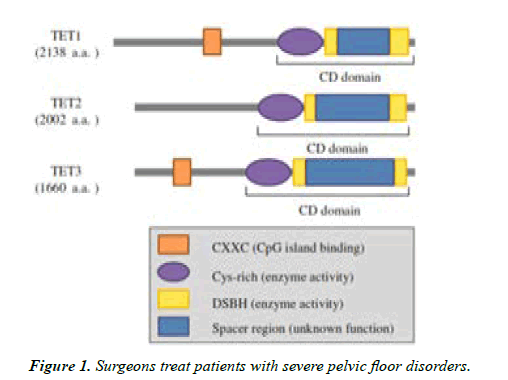
In the center of TET protein genes, exactly before the doublestranded β-helix, a cysteine rich region with unidentified function exists. The N-terminal area of TET1 and TET3 maintains the evolutionarily preserved CXXC-type domain which helps as a vital recognition in binding to un methylated cytosine moieties of DNA. Unlike TET1 and TET3, TET2 does not contain a CXXC-type domain [15,16] (Figure 1).
The C-terminal central part catalytic domain found in all TET proteins consists of the DSBH, a cysteine-rich domain. The DSBH domain has a huge low complexity part varying in length along with family of TET proteins. TET1 and TET3 contain N-terminal CXXC domain that can bind directly to unmethylated region of DNA and assist in marking genomic target site [13].
DNA methylation
Epigenetic is defined as the study of a gene function which can be passed from one cell to the next cell generation without an involvement of alteration in the DNA base sequence. DNA methylation and histone modi?cation are the two major mechanisms that represent epigenetic events [17-20]. Our review principally focuses on the mechanism of DNA methylation, which catalysed by a family of three DNA methyl transferase enzymes (DNMTs): DNMT1, DNMT3A and DNMT3B. DNA methyl transferase 1, the primary enzyme, is considered a continuance methyl transferase, for the reason that it is responsible for conveying methylation pattern from one cellular generation to the next [21-23]. Human genomes encompass comparatively equivalent proportions of the four DNA nitrogenous bases (A, G, C, and T). In addition to their original states, some of these bases can also be undergo reversible modifications; the most known one is being addition of -CH3 to cytosine base [11].
Cytosine methylation in DNA is a common covalent modification wherein a methyl group (-CH3) is added to the aromatic ring of a cytosine base (5-mC) within a CpG dinucleotide identified by DNA methyl transferase enzymes. The unmethylated CpG di nucleotides are highly concentrated in CpG rich regions, known as CpG islands (CGIs), sequences of consisting of about 1 kb in length. In the CpG islands, the CG content accounts for greater than 55% that is typically associated to the promoter regions and to the first non-coding regions (exons) of about 60% of genes [23,24].
CpG islands are evolutionarily conserved region of DNA that enhance gene expression by controlling the chromatin arrangement and transcription factor binding, thus, methylation of CpGIs is able to impair binding of transcription factor, recruit repressive methyl-binding proteins, and firmly silence gene expression [1].
The mechanism responsible for CGIs hypo methylation throughout the period of overall de novo methylation at early development remains unclear yet. However, some possible mechanisms were proposed which involve:
(i) CGIs remain hypo methylated is related to intrinsic sequence property which can exclude the activity or association of DNA methyl transferase.
(ii) CGIs attain DNA methylation generally, but are targeted by a demethylating action.
(iii) The basic transcriptional apparatus (RNA Pol II and TF) and histone H3 lysine 4 trimethylation (H3K4me3) prohibits the DNMTs from transcriptional initiation sites [25]. All of the three identified DNA methyl transferase are proteins with multi domains encompassing a large N- terminal region and a smaller C-terminal domain harbouring the catalytic core. DNMT1 shows a partiality for hemi-methylated DNA more than unmethylated in human cells [26]. Methylation occurs at cytosine residue that precedes a Guanosine in the CpG dinucleotide of DNA. Unmethylated CpGs are not arbitrarily distributed within the DNA sequence, however, are often gather together in ‘CpG islands that are in the promoter region of several genes [9,20,26] (Figure 2).
DNA methylation pathways
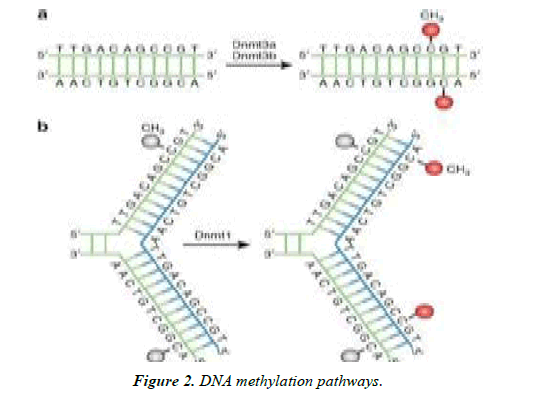
(a) DNMT3a and DNMT3b represent the de novo DNMTs and transfer methyl groups (red) from methyl group donor, S-adenyl methionine (SAM) onto the 5’ carbon of cytosine residue of bare DNA to form 5mC.
(b) DNMT1 is preservative DNMT and maintaining DNA methylation pattern at DNA replication level. When the DNA molecule undergoes semi-conservative replication, the parental DNA template maintains the original DNA methylation model (gray), [1].
DNA Methylation and control of Gene Expression
DNA methylation is a steady process; however, reversible covalent modifications that may change the arrangement of chromatin structure and gene regulation. Various organisms use this type of DNA modifications to regulate transcriptional process [27,28]. The status of DNA methylation affects several biological processes during mammalian developmental stages and is well-known to be enormously aberrant in cancerous cells [29]. DNA methylation influences activity of gene directly through inhibition of transcription factors binding and indirectly by employing chromatin associated proteins with suppressive properties [1,3,9,29]. In addition, it contributes to cell lineage restriction and genetic imprinting, for instance silencing of the X-chromosome in female mammals, during developmental period [22,30,31].
Methylation of 5’-carbon of cytosine in the CpG dinucleotide sites is an evolutionarily conserved epigenetic modification which partakes in regulation of gene expression without alterations in basic DNA sequences [32,33]. For example, the loss of DNA-methyltransferase1 activity, well demonstrated event in knockout mice, results in the abnormal differentiation of hematopoietic stem cells (HSCs) into myeloid progenitor cells (MPCs), [19] (Figure 3).
DNA methylation and regulation of gene expression
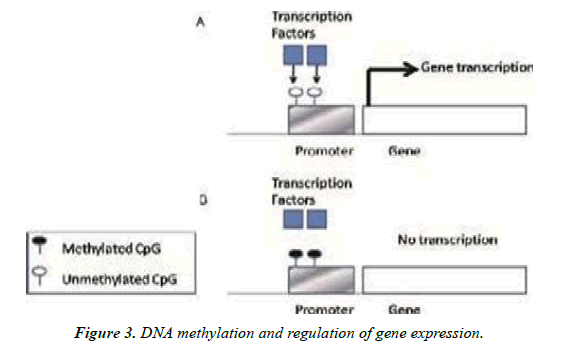
(A) Unmethylated CpG island promoter let’s binding of transcriptional factors, which is necessary for initiation of transcription.
(B) The methylated CpG island promoter keeps away transcriptional factors from binding of and leads to gene silencing [9].
The role of TET proteins in regulation of DNA methylation
Until the finding of TET1, DNA methylation was supposed to be an irreversible epigenetic change associated with gene repression, which might merely be assuaged through DNA replication [34]. The discovery of the Ten-Eleven Translocations (TET), hydroxylases that catalysed DNA demethylation has basically changed the conception of how DNA methylation is regulated [35]. Consequently, the discovery of TET family proteins capable of oxidizing 5mC to 5hmc, 5Fc and 5caC in DNA molecule has lead to deep progress in perception of the basic mechanism underlying DNA demethylation [2,4,32,36]. The TET proteins, DNA demethylases, actively down regulate the DNA methylation level. On the one hand, TET proteins regulate DNA methylation through binding to CpG-rich regions of DNA to prevent unnecessary DNA methyl transferase activity; on the other hand, they regulate DNA methylation by changing 5mC to 5hmC via their hydroxylase activities [14].
Several studies have suggested that there might be multiple mechanisms by which 5hmC and TET proteins regulate DNA methylation and gene expression processes [15]. The conversion of 5mC to 5hmC by TET proteins, leads to failure of DNA methylation during cell division. The derivative of 5mC that are produced by TET proteins are probable to serve like intermediates in DNA demethylation pathway. So far, two principal DNA demethylation processes have been proposed: The passive and active DNA demethylation pathways [7,18,37].
The Passive DNA Demethylation Pathway
The passive DNA demethylation pathway refers to the failure to retain DNA methylation pattern across cell divisions, and is supposed to be the outcome of replication dependent 5-methylcytosine dilution. Beside down regulating of the DNA methylation machinery, the deposition of 5-methyl cytosine can have a function in provoking passive DNA demethylation. The passive loss of 5mC can hence be accomplished through consecutive cycles of DNA replication in the absence of functional DNMT1/UHRF1, passive dilution of 5mC, [34,38]. The passive demethylation most probably occurs during mammalian developmental stage and cell differentiation, mostly in the maternal genome at some stages of pre implantation growth [18].
Conversion of 5-metlylcytosine to 5-hydroxymethylcytosine might promote passive demethylation as a result of replication by inhibiting the activity of DNMT1 to distinguish 5hmCpGs region as observed in vitro experiments. The TET-mediate 5hmC deposition may thus elicit passive replication dependent DNA demethylation on the opposite DNA template and be essential to counteract irregular assembly of atypically methylated pattern. However, the compel DNMT1 collaborator UHRF1 has been reported to bind 5-mC and 5-hmC with comparable affinities, suggesting that further investigation is required [7].
The active DNA demethylation
Active DNA demethylation is defined as an enzymatic reaction in which 5-mC base, in its oxidized form, is substituted by unmodified cytosine in a replication- independent mode [35]. In the active DNA demethylation pathway, multi enzymes including TET1, TET2 and TET3, which catalyse the conversion of 5mC to 5hmC, thymine DNA glycosylate (TDG), and methyl-CpG-binding domain protein 4 (MBD4), the RNA/DNA editing enzyme activation induced cytidine deaminize (AID), and growth restrain and DNA damage 45 (GADD45a, b, and c), which employs nucleotide and /or base excision repair (BER) factors to gene specific loci and serve as adapter molecules between the repair factors and chromatin are involved [18,39].
A number of mechanisms regarding to active DNA demethylation pathways have been hypothesized; however, base excision and DNA base excision repair (BER) catalyzed by thymine–DNA–glycosylase (TDG) are currently assumed to be the major imposer. Thymine DNA glycosylase is a DNA mismatch repair enzyme which binds and excises impaired pyrimidine’s in G:U and G:T base pairs [35,39]. The DNA mismatch repair enzyme, TDG, was initially identified as recognizing and repairing T:G mismatches in DNA base pairing. However, TDG also capably excises 5fC and 5caC that are correctly paired to G in double strand DNA, and depletion of TDG leads to increased levels of 5fC and 5caC at a particular gene location [7,34].
Biochemically, the TET catalyzed DNA demethylation pathway should be irreversible, because no replication dependent means is found to resynthesize 5mC oxidation products. However, note that, the levels of both 5hmC and 5mC are comparatively stable in normal mammalian tissues [36]. Most evidences exist for mechanisms by which TET proteins convert 5mC into 5hmC, 5fC and 5caC through three successive oxidation reactions. Furthermore, evidence exists for a mechanism by which AID/APOBEC proteins delaminate 5hmC to 5hmU followed by TDG mediated base excision repairs [37].
Expression TET proteins in different tissues
All TET family members possess the ability to oxidize 5mC to 5hmC, and 5fC5cac, however, they are not equally expressed in different cells or tissues. For instance, the expression of TET1 and TET2 is mostly occurred in mouse embryonic stem cell (ESC), while TET3 is mostly expressed in oocytes and one cell zygotes [7,13]. Even though they were recognized only a few years ago, increasing evidence indicates that TET proteins have critical roles in epigenetic modification of stem cell differentiation, development of embryo, growth, and tumor cell formation [9].
On the contrary to the association of 5mC oxidation with regional restoration of an unmethylated form during reprogramming, in somatic cells like stem cells and neuron cells, 5mC oxidation usually appears to contribute to the continuance of methylation fidelity by counteracting aberrant de novo methylation process. In embryonic stem cells, TET1 keeps CpG-rich promotes hypo methylated by getting rid of erroneously placed methylation [3]. Existing evidence suggested that oxidation of 5-mC in the paternal genome in fertilized eggs by TET3 starts DNA demethylation and stimulates the activation of the paternal replica in the early embryonic genes [7].
Physiological functions of TET proteins
The physiological importances of the TET proteins have been investigated using genetic knockout mouse models. Constituent deletion of TET3 results in neonatal lethality with 100% penetrance, while the deletions of either TET1 or TET2 do not result in any noticeable developmental phenotype. This implies that TET3 has an exceptional role during embryogenesis that cannot be remunerated for by other TET enzyme family [34].
Mechanisms of TET protein regulation
The demonstration of Bauer et al. revealed that TET proteins are subjected to diversity of post-translational modifications that typically occur at specific regulatory regions (N- terminus and low-complexity regions found between the domains of di oxygenase). Their results provided a new possible means for TET protein regulation based on a dynamic interchange of phosphorylation and O-linked GlcNAcylation at specific regulatory regions [40]. Away from gene expression, the activity of TET is influenced by a variety of cellular processes, including alteration in levels of reactive oxygen species (ROS), Krebs cycle intermediates (succinate and fumarate) compete with TET for its cofactor α - ketoglutarate, and proliferation [41].
Many proteins including components of chromatin, subcellular localization and chromatin-binding interact with TET proteins and involved in the regulation of their activities. Additionally, many other molecules were also identified to be involved in the regulation of TET enzyme activity. Ascorbic acid (vitamin C) and ATP, which not directly participate in the hydroxylation of 5hmC are the two most common molecules demonstrated both in vitro and cultured cell to have triggering effect on enzymatic activity of TET proteins [3,40]. However, in 2013, Yin et al. confirmed that ascorbic acid can directly involve in the regulation of TET proteins by enhancing their activity through unclear mechanism [36].
TET Proteins and Cancer
DNA methylation pathways has appeared to be an important epigenetic event that plays crucial roles in cell growth, apoptosis, aging and diseases such as cancer [42]. The elevated rate of mutagenesis of 5mC compared to other nucleotides has been broadly documented in the literatures. The mutation rate of CpG di nucleotides has been predicted to be about 10 to 40-fold compared to other dinucleotide mutation rate [21]. Many research results have also showed 5hmC as a potential biomarker whose reduction is associated with tumor development and spread [42].
Changes in DNA methylation model have been reported in several malignant cells and revealed to play a vital role in cancer differentiation and progression processes [33,43]. The balance between DNA methylation and demethylation pathways is controlled by many proteins and cofactors. In the case of cancer, this balance is recurrently deregulated and leads to altered DNA methylation patterns, which in turn results in the repression of tumor suppressor gene or activation oncogene [37]. As the loss of 5mC is a feature of nearly all cancer types, active DNA demethylation pathway plays a significant role in the preservation of DNA methylation patterns, and perhaps, of other epigenetic change mark [43].
The TET proteins whose name was first derived from the TET1characterization as a fusing partner in mixed- lineage leukaemia (MLL), which has been identified in both lymphoid and myeloid cancer cases [13]. The most common cytogenetic findings of acute myeloid leukaemia (AML) showed aberrations of chromosomal gene involving the MLL at 11q23. The (t (10, 11) (q22; q23)) has been frequently reported in several cases of AML [43]. In 2009, TET2 aberration was found in various haematological malignancies. The TET2 gene was found to c be affected by deletion, splice site, and point mutations such as missense and nonsense [37]. In 2013, Asmara and his colleagues found that TET2 mutations in 12% of diffuse large B-cell lymphoma (DLBCL), with 5% carrying missense and 7% carrying nonsense, frame-shift or splice-site mutations with a mutation detecting analysis focused on TET2 gene [35].
The gain of function of oncogene in IDH mutation is an initiation event that identifies main clinical and prognostic classifications of gliomas. Mutant IDH proteins produce a new on co-metabolite, 2-hydroxyglutarate that competes with a-ketoglutarate for active site of the DNA modifying enzymes of the TET family and interrupts TET function [44,45].
Conclusions and Perspectives
In summary, the basic concepts regarding to the structure and the regulation role of the TET proteins in DNA methylation as well as in development of cancers have been covered in this review article. Several studies revealed that TET proteins play a crucial regulatory role in DNA methylation. The TET proteins, DNA methyl transferase, catalyze DNA methylation through reversible covalent biochemical modification by addition of a methyl group (-CH3) to the aromatic ring of cytosine residues (5mC) within a CpG di nucleotides. Nowadays, the discovery of the TET proteins has dramatically changed the assumption that belief the DNA methylation to be an irreversible epigenetic event associated with gene suppression and revealed the mechanisms by which they regulate DNA methylation.
In addition regulation of DNA methylation, recent findings showed that TET proteins play crucial roles in epigenetic remodelling in stem cell differentiation, embryogenesis and control of cancers. In general, since TET proteins are the key regulators of DNA demethylation pathways and gene expression, the mutation in the TET protein genes or the dysregulation of their activities or alteration in their expression levels can lead to the development of different cancers. Therefore, the future research findings should focus on how to target TET proteins as potential biomarkers of tumor cells at early development in order to control cancers that are developed as a result of TET proteins mutation.
Authors’ contribution
Teka Obsa conceived and designed the review, searched organized and interpreted journal articles and wrote the manuscript. Mezgebu Legesse, Getahun Chala, Mohammed Yusuf, Tefera Belsty and Wubshet Nebiyu Searched and organized and interpreted journal articles and wrote the manuscript. All the authors read, commented on, and contributed to the submitted and revised manuscript.
Declaration of interest
The authors have no conflicts of interest to declare.
References
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuro psycho pharmacol. 2013;38(1):23-38.
- Shen L, Zhang Y. Enzymatic analysis of Tet proteins: key enzymes in the metabolism of DNA methylation. In Methods in enzymol. 2012;512:93-105.
- Xu GL, Walsh CP. Enzymatic DNA oxidation: mechanisms and biological significance. BMB reports. 2014;47(11):609.
- Ko M, An J, Pastor WA, et al. TET proteins and 5?methylcytosine oxidation in hematological cancers. Immunol rev. 2015;263(1):6-21.
- Delatte B, Fuks F. TET proteins: on the frenetic hunt for new cytosine modifications. Briefings in Functional Genomics. 2013;12(3):191-204.
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol cell biol. 2013;14(6):341-56.
- Tsagaratou A, Rao A. TET proteins and 5-methylcytosine oxidation in the immune system. InCold Spring Harbor symposia on Quantitative Biol. 2013;78:1-10.
- Wang Y, Zhang Y. Regulation of TET protein stability by calpains. Cell Rep. 2014;6(2):278-84.
- Lim DH, Maher ER. DNA methylation: a form of epigenetic control of gene expression. The Obstetrician & Gynaecol. 2010;12(1):37-42.
- Majerski AA, Quinton AC, Marsden PA. Epigenetic Mechanisms of the Vascular Endothelium. InEpigenetics and Epigenomics. 2014.
- Teperino R, Lempradl A, Pospisilik JA. Bridging epigenomics and complex disease: the basics. Cellular Mol Life Sci. 2013;70(9):1609-21.
- Zhu H, Wang G, Qian J. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genetics. 2016;17(9):551-65.
- Chen HF, Wu KJ. Epigenetics, TET proteins, and hypoxia in epithelial-mesenchymal transition and tumorigenesis. Biomed. 2016;6(1):1-8.
- Li D, Guo B, Wu H, et al. TET family of dioxygenases: crucial roles and underlying mechanisms. Cytogenetic and Genome Res. 2015;146(3):171-80.
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Develop. 2012;139(11):1895-902.
- Nakajima H, Kunimoto H. TET2 as an epigenetic master regulator for normal and malignant hematopoiesis. Cancer Sci. 2014;105(9):1093-9.
- Kirchner H, Osler ME, Krook A, et al. Epigenetic flexibility in metabolic regulation: disease cause and prevention?. Trends Cell Biol. 2013;23(5):203-9.
- Li CJ. DNA demethylation pathways: recent insights. Genetics and Epigenetics. 2013;5:GEG-S12143.
- Martino D, Kesper DA, Amarasekera M, et al. Epigenetics in immune development and in allergic and autoimmune diseases. J Reproductive Immunol. 2014;104:43-8.
- Zeisberg EM, Zeisberg M. The role of promoter hypermethylation in fibroblast activation and fibrogenesis. J Pathol. 2013;229(2):264-73.
- Akhavan-Niaki H, Samadani AA. DNA methylation and cancer development: molecular mechanism. Cell Biochemistry Biophysics. 2013;67(2):501-13.
- Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12(1):1-3.
- D’Aquila P, Rose G, Bellizzi D, et al. Epigenetics and aging. Maturitas. 2013;74(2):130-6.
- Chiacchiera F, Piunti A, Pasini D. Epigenetic methylations and their connections with metabolism. Cellular Mol Life Sci. 2013;70(9):1495-508.
- Illingworth RS, Bird AP. CpG islands–‘a rough guide’. FEBS letters. 2009;583(11):1713-20.
- Siddique AN, Nunna S, Rajavelu A, et al. Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a–Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol. 2013;425(3):479-91.
- Ikeda Y, Nishimura T. The role of DNA methylation in transposable element silencing and genomic imprinting. In Nuclear Functions Plant Transcription, Signaling Develop. 2015;13-29.
- Plongthongkum N, Diep DH, Zhang K. Advances in the profiling of DNA modifications: cytosine methylation and beyond. Nat Rev Genetics. 2014;15(10):647-61.
- Koh KP, Yabuuchi A, Rao S, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell stem cell. 2011;8(2):200-13.
- Gut P, Verdin E. The nexus of chromatin regulation and intermediary metabolism. Nat. 2013;502(7472):489-98.
- Guo P, Yan S, Hu J, et al. Selective detection of 5-formyl-2′-deoxycytidine in DNA using a fluorogenic hydroxylamine reagent. Organic letters. 2013;15(13):3266-9.
- Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell biol. 2015;16(10):593-610.
- Lubecka-Pietruszewska K, Kaufman-Szymczyk A, Stefanska B, et al. Folic acid enforces DNA methylation-mediated transcriptional silencing of PTEN, APC and RARbeta2 tumour suppressor genes in breast cancer. Biochem Biophys Res Communicat. 2013;430(2):623-8.
- Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes & Develop. 2016;30(7):733-50.
- Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematol. 2013;98(12):1912.
- Yin R, Mao SQ, Zhao B, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J Am Chem Soc. 2013;135(28):10396-403.
- Kroeze LI, van der Reijden BA, Jansen JH. 5-Hydroxymethylcytosine: an epigenetic mark frequently deregulated in cancer. Biochimica et Biophysica Acta (BBA)-Rev Cancer. 2015;1855(2):144-54.
- Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156(1-2):45-68.
- Li X, Wei W, Ratnu VS, et al. On the potential role of active DNA demethylation in establishing epigenetic states associated with neural plasticity and memory. Neurobiol learning memory. 2013;105:125-32.
- Bauer C, Göbel K, Nagaraj N, et al. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT). J Biol Chem. 2015;290(8):4801-12.
- Thienpont B, Steinbacher J, Zhao H, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nat. 2016;537(7618):63-8.
- Gambichler T, Sand M, Skrygan M. Loss of 5-hydroxymethylcytosine and ten-eleven translocation 2 protein expression in malignant melanoma. Melanoma Res. 2013;23(3):218-20.
- Plass C, Pfister SM, Lindroth AM, et al. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genetics. 2013;14(11):765-80.
- Carroll MP. When cancer and immunology meet. Immunol Rev. 2015;263(1):2.
- Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nat. 2016;529(7584):110-4.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref