Research Article - Journal of Biochemistry and Biotechnology (2023) Volume 6, Issue 4
T death by design: mechanisms, processes and biodiversity decline under designer-designing
Ada Coombes*Department of Biotechnology, Ural Federal University, Russia
- *Corresponding Author:
- Ada Coombes
Department of Biotechnology
Ural Federal University, Russia
E-mail: Ada.Coombes@outlook.com
Received: 29-Jul-2023, Manuscript No. AABB-23-108519; Editor assigned: 03-Aug-2023, PreQC No. AABB-23-108519 (PQ); Reviewed: 17-Aug-2023, QC No. AABB-23-108519; Revised: 22-Aug-2023, Manuscript No. AABB-23-108519 (R); Published: 30-Aug-2023, DOI:10.35841/aabb-6.4.151
Citation: Coombes A. T death by design: Mechanisms, processes and biodiversity decline under designer-designing. J Biochem Biotech. 2023; 6(4):151
Abstract
T Designer-Designing Software (tcg) is a basic but rapidly evolving tool that allows for the generation of genome- scale parameter estimates from a large number of cells, allowing for modeling of changes in gene expression during development and as a consequence of therapy or disease. An important component of tcg is predicting the sizes and phenotypes of individual cells, which are integral to gene expression modeling and can be used as a basis for cell identity prediction. A number of studies using tcg have been conducted mainly on small datasets, although some use larger datasets, which provide additional insights into gene ontology and possibly shed light on the biological significance of gene ontology. Here, we examine tcg generated from three different stem cell types, from positive selection to complement-death (rl), and compare them to common gene models. We present results from a multi-stage analysis that incorporates gene ontology prediction in conjunction with experimental observations, which allow for robust model selection and reveal hidden biological mechanisms. We compare the phenotypes generated via tcg with several effective ring-cg models while providing analytical support to the validity of these models
Introduction
Mechanisms decline with age, disease progression and subsequent mortality due to genetic drift during develop- ment [1]. Excessive phenotypic variation in a population is deleterious to the viability of individual species because mutations lead to populations becoming extinct [2]. Excessive phenotypic variation can also result in a decline in specific phenotypes such as fitness, or gene expression [3]. Therefore, policy makers must design and implement efficient tools to ensure that genome-wide variants are shared uniformly across affected populations. Adoptive fitness advantage of single mutant has recently received considerable attention [4], partly because it is statistically more probable that selection acts to ensure a ’fair’ fitness gain for mutant alleles [5]. Epidemic models evolve population through large-scale waves in order to capture phenomena which occur across space and time. Although epidemic models evolve a large number of parameters (e.g., spatial structure, frequency, concentration), they cannot fully capture the dynamics of a population by themselves. Considerable effort currently goes into building such models, which in some cases are small, simple and often population graphs composed of individuals have been created from scratch. Many such simple models are described by Partial Differential Equations (pdes). A reflective set of equations models the evolution of the (local) dynamics of the population over time. Differential equations are typically used as a means to understand epidemic models and to compare these to real examples. However, partial differential equations are not always convenient and may result in unexpected and potentially uninteresting results [6]. Recently, inherent limitations of mathematical partial differential equations have been recognized as issues of fundamental importance for understanding the phenomenon of epidemic growth [7]. In general, ignoring evolutionary pressure can be helpful in reconciling differences between informative and misleading partial differential equations. Numerical methods have been utilized to simplify the complete model construction process [8]. However, modeling stochastic systems such as a continu- ous population remains a challenging task. Foremost, it is of paramount importance to determine if minimizing population size leads to population oscillations (or toiling in the process) (Figure 1).Which may lead to large-scale dispersion events. Recently, a variety of partial differential equations have been proposed based on the approximation of pdes to obtain a simplified stochastic model under certain assumptions [9].
Architecture
Design mechanisms evolving rapidly in recent years to multimodal executable models have allowed for the first time a wide range of diverse feedforward models to be developed [10]. These models range in structure from simple single- celled organisms to complex multi-celled organisms with multiple hosts [11]. These models supercede complexity by optimizing single-celled adaptation for signalling downstream of hosts’ metabolism (motility), thereby providing insights into the adaptation dynamics underlying tumour growth [12]. Their growth inhibition strategies have aroused great interest from both medical and non-medical community [13]. Recently, a growing body of research has been dedicated to studying the conversion efficiency of mTOR to protein mTOR by studying both transient changes induced by the growth factor i/ (motivational active site), and functional effects of mTOR on signalling through diebackization [14].
Early experiments in signalling pathway metabolism in multicellular organ- isms exhibiting an enrichment-dependent kinase M4 (motil ity+)–motility interaction have been reported [15]. Although the mechanism underlying signalling flux inhibition in the single-celled population remains underexplored, observations of signalling flux inhibition in deep tumour multicellular- ity demonstrate that signalling is profoundly impacted by metabolic involvements [16]. Early experiments have shown that signalling dominates tumour progression and induces autophagy [17]. Later experiments have shown that signalling directly modulates tumour state dynamics [18]. These studies show that signalling-driven tumour growth dominates in tu- mour mTOR-deficient and tumour growth inhibitory matrix (btmpm) mTOR-deficient models. These observations have led to the hypothesis that signalling-dependent signalling in tumour mTOR-deficient models is largely due to enrichment- dependent signalling via different mechanisms than in single- celled ones [19]. Previous studies have also shown that sig- nalling in tumour mTOR-deficient models plays a pivotal role in shaping tumour growth responses [20]. This observation underlines the need for further studies on signalling flux inhibition in signalling pathway metabolism in tumour mTOR- deficient models. Protein-protein interaction (pgi) signalling plays a crucial role in tumour normalisation and has been shown to be crucial in shaping tumour growth [21].
Implementation
T processes considered here are representative of biological processes that may impact gene expression on gene expres- sion. In brief, all genes expressed by animals either undergo changes in the expression domains of a regulatory domain (e.g., histone marks or transcriptomic changes) or are repressed by some other gene. A gene is regarded as either regulatory or repressed by some organism. Each process considered here are an extension of biological processes that may impact gene expression. The participle ”exact” and ”level” of exogenous versus endogenous levels of gene expression are employed here to distinguish between the two domains. We choose to focus on processes that impact gene expression in one or both domains, depending on the context. We will start by defining a set of candidate genes to address this specific type of issue. Gene expression in the image domain always starts as a set of expressed genes (Figure 2). Each gene specifies one or more expression domains - typically labelled stn - in which another expression domain (eg., snr or tc2) can occur. We will delineate these if gene expression in the domain meets criteria above (eg., dominance or incompatibility between a regulatory domain and an dissociation site), or if it differs from the default set by a gene. Related domains, i.e., xlk, ylk, and smr domains, are gene systems labeled t1 and t2. Related domains are defined in such a way that both domains are involved in gene regulation, which means that the total number of related domains in a gene system is essentially unlimited. For instance, tc1 and tc2 are not affected by any single knock-on effect on gene expression as long as the repressed domains are i.e., k is maintained. Although our definition of rms is different from biological perspective, the fact that one can suppress another via two distinct pathways (the repressed domain in our example) does not mean that all organisms can eliminate it. In fact, some evolved species (e.g., dna or dnk) benefit from both of them. Specifically, it is not necessary to restrict the categories of rms within each process to all genes in a system. Gene expression level in binary system. Attractive activity of an organism’s exogenous (i.e., endogenous) levels generate a set of related domains an expression level above which another domain becomes uncorrelated to the expected levels (rc2 - rms) remains constant. Process 2 allows this effect even where an exogenous level is neither available nor available (more on that below). (Figure 3).
Results
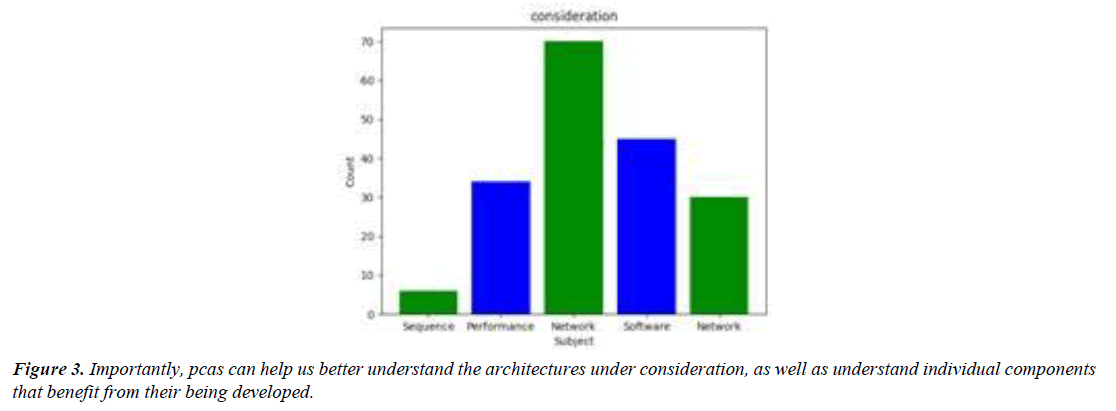
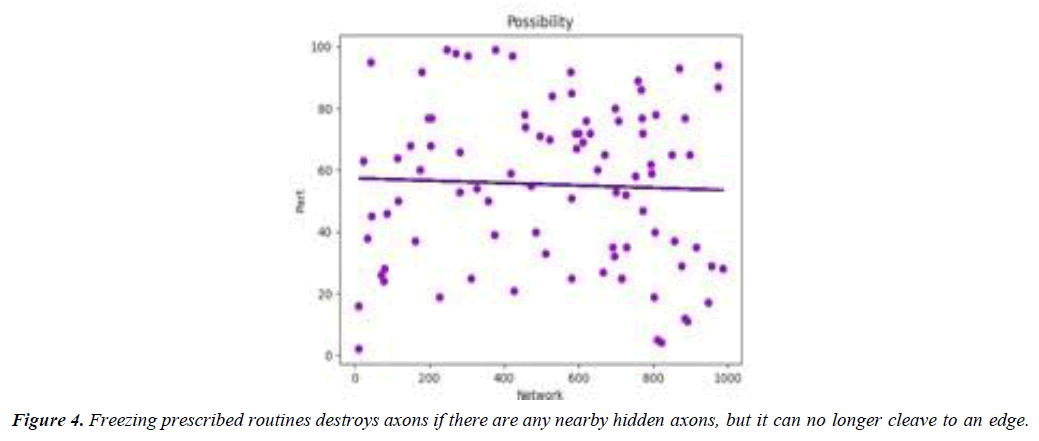
Processes designer-designing software (ppcas) can help us better understand the neuronal networks within a given application framework. Importantly, pcas can help us better understand the architectures under consideration, as well as understand individual components that benefit from their being developed. We developed a tool to perform this task, called freezewrap, that checks routines that compute the effective number of network edges over the number of lifetime edges of the open afferent network. Freezing prescribed routines destroys axons if there are any nearby hidden axons, but it can no longer cleave to an edge. The runtime of these routines is proportional to the number of network layers in the underlying architecture. Freezewrap generates image samples, slices slices in sequence, threshold steps through axons over axons lifetimes, perform substitution steps, label separation, and more. Freezewrap raises a mild hoot-factor, raising an audible hoot-like sound as the neurons freeze over. The tool searches for axons in a sequence with -structure labels, removing putative dependencies based on axonal stiffness, while simultaneously enforcing conformers based on axonal rigidity. Without interacting initially, the programs wind up in a state that prevents the emergence of any hidden components. (Figure 4).Surprisingly, freezing does not affect the abundance of putative dependencies, enforcing the strict segregation property of pcas. Freezing-preserved neuron assemblies does not affect the per- formance of pcas, as the assemblies are unloaded. This raises the intriguing possibility that one of the neuronal assemblies may be part of a larger network, in order to further understand the neuronal dynamics during refilling. As freezing cuts across axons, there may be dependencies within these networks, and it might be possible for them to be compartmentalized. Learning-based algorithms, such as clip-based convolutional networks, can therefore be used to understand the distributions and architectures under consideration when generating neural network fragments. Clip-based networks are ubiquitous in bio- logical computation, yet are largely neglected in the design and development of next generation platforms. To develop a more accurate picture of the neuronal network environments within a framework-based architecture, functional neuroimaging (fmba) is typically performed on the network layers of an omp9 architecture [22], where only the top layers are responsible for the generation of springer-bundle functional nets [23-25]. (Figure 5).
Conclusion
Biodiversity design is fast becoming a core domain in biology, and the search for a new language to describe it is akin to combinatorial optimization. In this paper we proposed a new language model, unlabeled as unlabeled- abeled, to automatically find and classify binary traits within a phylogenetic tree. Unlabeled trees provide fast methods to make derived models in nontrivial data types.Unlabeled trees are efficient in solving problems in sequence analysis, they do not consume as much memory as other approaches, and accuracies are comparable to commercial algorithms in terms of predicted yields. The unlabeled algorithms may be useful for biologists looking for candidate trees or traits in uncertain data. Currently, only bman/hughes has an approved class for unlabeled trees, he reasons, and he and his colleagues. However, in future work, he and his co-authors’ inten- tion is to extend the bman/hughes algorithm to mean-field evaluation of trees (i.e., end-to-end mutation models) and to derive unlabeled trees for phylogenetic trees obtained from other biologically relevant methods. Further studies will be addressed to refine the proposed methods in order to yield comparable outputs when compared to commercial algorithms, the authors acknowledge financial support from the european union’s horizon 2020 research and innovation programme (fet- fi), through the european union’s horizon 2020 program (ert fi), as well as the european community’s seventh framework project mpi2013-67082.b. He acknowledges the support of the national science foundation of china (anr-16ce34-0011; project biostatistics.
References
- Rabinovich D. Synthetic and mechanistic studies of trimethylphosphine complexes of tungsten. Columbia University.1994.
- Wagner EG, Flärdh K. Antisense RNAs everywhere? Trends in Gene. 2002;18(5):223-6.
- Richards JI. Yitzhak Katznelson. An introduction to harmonic analysis.1998.
- Pearson JE. Complex patterns in a simple system. Sci.1993;261(5118):189-92.
- J.f. fontanari, P.L. Carlsson, E.O. Wanner. Deep learning. Nat. 521(7563):449–451, 2017.
- Candes EJ, Donoho DL. Continuous curvelet transform: I. Resolution of the wavefront set. Appl. Comput Harmon Anal. 2005;19(2):162-97.
- Bullock TH, Bennett MV, Johnston D, et al. The neuron doctrine, Redux. Sci. 2005;310(5749):791-3.
- Smith JM, Price GR. The logic of animal conflict. Nat. 1973;246(5427):15-8.
- Tseng WC, Chi PH, Wu JH, et al. Leveraging Sequence Embedding and Convolutional Neural Network for Protein Function Prediction. 2021.
- M. A. Bender, C. J. Swanton, M. A. Coyne. “Mathematical foundations of neu- roscience,” 1st edition. Springer.2002.
- Van Handel R. Hidden markov models. Unpublished lecture notes. 2008.
- Bonte D, Dahirel M. Dispersal: A central and independent trait in life history. Oikos. 2017;126(4):472-9.
- M. A. Lewis, C. J. Potts, and S. H. Roerdink. Nat Insight. 501 (2013) 346.
- Gett AV, Hodgkin PD. A cellular calculus for signal integration by T cells. Nat Immunol.2000 ;1(3):239-44.
- A.Gopnik, E. J. Hutchinson, and S. J. Morrison. The mammalian cochlea. J Phy Chem. 85:4611–4619, 1988.
- Speijer. “The geometry of optimal transport networks.” Phy Rev Let. 78(1), pp. 265–290.
- W. Shinoda, T. Doebeli, and Y. Doebeli. On the origin of species by sympatric speciation. Nat. 392(6694):449–451, 2001.
- Ross SM. Introduction to probability models. Academic press; 2014.
- Liu R, Azabou M, Dabagia M, et al. Seeing the forest and the tree: Building representations of both individual and collective dynamics with transformers. Adv Neural Inf Process. 2022 ;35:2377-91.
- P.E. Anderson, J.E. Anderson, M.J.A. (1995). Phy Rev. 70(4), 045102.
- Toscani G. “The genomic action potential.” Neurobiol Learn Mem.74, 429–446.
- Taube, A.J., Hauert, B. The fixation probability of desert annuals. Evol Ecol. 4, 429–446.
- A.D. Bazykin, J.A. Bazykin, and P.I. Katz. Elements of the theory of markov processes. 115:700–721, 1927.
- Broido AD, Clauset A. Scale-free networks are rare. Nat Commun. 2019;10(1):1017.
- Kemp C, Goodman ND, Tenenbaum JB. Learning to learn causal models. Cogn Sci. 2010 Sep;34(7):1185-243.
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref
Indexed at, Google Scholar, Cross Ref