Research Article - Biomedical Research (2017) Volume 28, Issue 15
Study on G-protein-coupled receptor polymorphisms reveals close relationship with asthma in a population of China
Zhen-gang Yu1,2 and Zhao-zhong Cheng3*
1Qingdao University, Shandong Province, Qingdao City, PR China
2Respiratory Medicine of Weifang People’s Hospital, Shandong Province, Weifang City, PR China
3Department of Respiratory, Affiliated Hospital of Qingdao University (Eastern Hospital), Qingdao City, Shandong Province, PR China
Accepted date: June 12, 2017
Abstract
To evaluate the relationship of G protein-coupled receptor for asthma susceptibility (GPRA) with asthma and the total IgE levels in a Chinese population. Six Single Nucleotide Polymorphisms (SNPs) from GPRA were determined by Taqman SNP genotyping assay and genotyped by studying the subjects of 143 cases and 171 controls. We assessed Hardy-Weinberg equilibrium in control group. Association between SNPs and corresponding haplotypes, IgE as well as asthma was studied. Regarding allelic distribution, current study showed similar pattern with that of other populations. For rs324981, the minor allele homozygotes AA lowered the risk for asthma (case-control OR: 0.34, P=0.002). rs324396 (TT) was protective for asthma (OR: 0.42, P=0.003). Although association between the six SNP haplotypes and asthma was not observed, haplotype H3 CTACGA was related to total IgE (OR: 1.53, P=0.004). Our study supports that GPRA gene plays a role in asthma susceptibility and also suggests two protective factors for asthma from this gene.
Keywords
Asthma, Single nucleotide polymorphism, GPRA, Immunoglobulin E, Chinese population
Introduction
Asthma is a severe episodic disease which features elevated responsiveness of the tracheobronchial tree, chronic inflammation and mucus secretion due to many stimuli [1]. Around 300 million people worldwide suffer from it with 250,000 deaths annually and the healthcare cost is mounting unprecedentedly [2]. With the rapidly accumulated knowledge on genetics and our current understanding of genetic differences which play an important role associated to the risk of asthmatic conditions, pinpointing those responsible specific genes as well as their Single Nucleotide Polymorphisms (SNPs) has been undertaken for the past several decades. Many candidate genes were proposed and clinically tested, including A disintegrin and metalloprotease33 (ADAM), dipeptidylpeptidase 10 (DPP10), inositol polyphosphate 4 phosphatase type I (INPP4A) and PDH finger protein 11 (PHF11) [3-6]. Since its discovery by genome-wide scan and positional cloning in a Finnish and a Canadian population [7,8], G protein-coupled receptor for asthma susceptibility (GPRA), also called G protein-coupled receptor 154 (GPR154) or Vasopressin Receptor-Related receptor 1 (VRR1) or human neuropeptide S receptor (NPSR1) [9] has been intensively studied with the majority of conclusions supporting the assumption that this gene was related to asthma pathogenesis [10,11]. This gene is localized on chromosome 7p with 7 SNPs and in a mouse model SNP-tagged haplotypes were also observed to be associated with high serum immunoglobulin E (IgE) or asthma [7]. Nevertheless, other studies suggested that this gene and its SNPs did not show participation in the occurrence of asthma and accompanying increased level of total IgE [12-14]. Contradicting reports require replication of studies with patients from different ethnics because one of the most important strategies in genetic research on sophisticated diseases calls for investigating new findings in populations with distinct origins and environmental conditions to support or defy the original hypothesis. In this study, we designed the case-control experiment on a population in Shandong province, China for the purpose of assessing the relationship between asthma, total serum IgE (tIgE) levels and GPRA.
Materials and Methods
Study population and IgE measurement
Based on the Global Initiative for Asthma criteria [15], asthma was diagnosed by a questionnaire to evaluate history of respiratory symptoms and physical conditions and the measurement of lung function of Forced Expiratory Volume in 1 s (FEV1) and Forced Vital Capacity (FVC). The recruited subjects met the following criteria: current physiciandiagnosed asthma, at least two or more symptoms, including episodic wheezing, breathlessness, cough and chest tightness, during the past 6 months. Controls were healthy residents in Shandong province, China with normal or near-normal lung function and no family or personal history of asthma. From the outpatient health centers in Shandong province we recruited 143 unrelated adult asthmatic patients (mean age 38.4 ± 13.2 y) and 171 controls (mean age 37.9 ± 11.7 y). Gender and age were matched between the asthmatic and control subjects. All subjects provided written consent after the detailed explanation by researchers. This study was granted by the Bioethics Committee of Qingdao University. Total concentration of IgE was quantified by ImmunoCAP total IgE kit (Phadia AB, Uppsala, Sweden).
Selection of single nucleotide polymorphism and genotyping
Laitinen et al. [7] initially reported 13 SNPs that tagged the most haplotypes related to asthma study. We selected five SNPs that are from intron 2 in GPRA: RS323917, RS323920, RS324377, RS324396 and RS324957. Moreover, one SNP in exon 3, rs324981, was also included because one study on a Chinese population reported the possible relationship between the disease and genotype [16]. Subsequently, all the selected SNPs were genotyped by using Taqman SNP genotyping assay provided by Applied Biosystems (CA, USA) and the quality value for automatic base-calling was above 99%. By referring to contig NT_007819.18, the chromosomal positions of studied SNPs are 34702031 (rs323917, C/G), 34707405 (rs323920, C/T), 34716363 (rs324377, A/C), 34750511 (rs324396, C/T), 34761760 (rs324957, A/G) and 34778501 (rs324981, A/T). The genomic DNA was extracted and purified from peripheral blood of the subjects adopting DNeasy Blood and Tissue Kit (Qiagen, Germany).
Statistical analyses
We applied Student’s t test and χ2 test to calculate the difference in distributions of quantitative and categorical variables between the groups. Each one of the SNPs in the control group was studied by Hardy-Weinberg equilibrium. We used Fisher’s exact test to detect allelic frequencies across the case and control group. For haplotypic analysis, Odds Ratio (OR) and Confidence Interval (CI) were calculated based on the frequency and estimated counts of all haplotypes contributed by each subject who provided two haplotypes from each chromosome. Statistical R package was utilized to perform all the statistical analyses needed [17].
Results
The general characteristics of the dataset (143 cases and 171 controls) are presented in Table 1.
| Items | Cases (N=143) | Controls (N=171) | P value |
|---|---|---|---|
| Age (means ± SD, y) | 38.4 ± 13.2 | 37.9 ± 11.7 | 0.319 |
| Male, n (%) | 61 (42.7) | 77 (45.0) | 0.275 |
| BMI (means ± SD, kg/m2) | 21.3 ± 3.6 | 22.1 ± 2.8 | 0.542 |
| FEV1 (L) | 2.38 ± 0.5 | 13.05 ± 0.47 | <0.05 |
| FVC (L) | 2.51 ± 0.93 | 3.48 ± 1.15 | <0.01 |
| Total IgE (means ± SD, IU/ml) | 429.4 ± 617.6 | 264.5 ± 502.4 | <0.01 |
Table 1. Demographic and clinical characteristics of the studied subjects from Shandong Province.
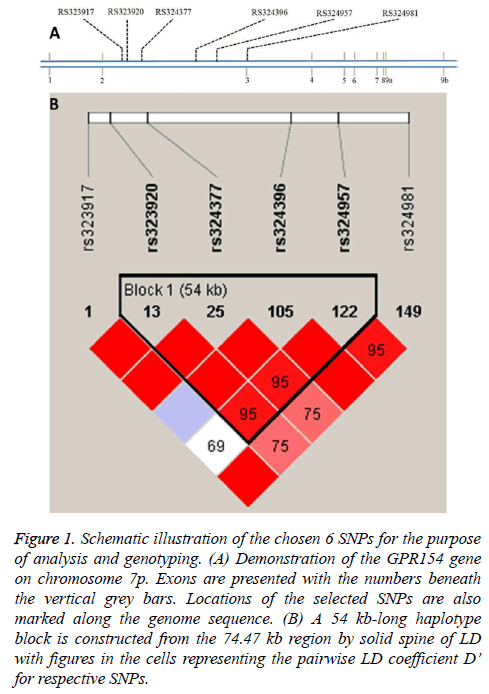
The subjects chosen were unrelated and no significant differences regarding age and gender as well as Body Mass Index (BMI) was found between the controls and cases. Figure 1a demonstrates locations on chromosome 7p for the six SNPs in this study. The SNPs were found to be in Hardy-Weinberg equilibrium (data not shown). From Figure 1b, variable pairwise D’ values were in the range from 0.69 to 1.0 and four SNPs were present in a 54 kb haplotype block by adopting the approach of solid spine of Linkage Disequilibrium (LD) [18]. In contrast, no blocks were detected with the algorithm of Gabriel et al. [19].
Figure 1: Schematic illustration of the chosen 6 SNPs for the purpose of analysis and genotyping. (A) Demonstration of the GPR154 gene on chromosome 7p. Exons are presented with the numbers beneath the vertical grey bars. Locations of the selected SNPs are also marked along the genome sequence. (B) A 54 kb-long haplotype block is constructed from the 74.47 kb region by solid spine of LD with figures in the cells representing the pairwise LD coefficient D’ for respective SNPs.
Data from the controls (Table 2) in Shandong Province indicated similar values to that reported in Canadian, Finnish, German and Chinese population [7,16,20]. Concerning cases, occurrence of cytosine in rs324396 and adenine in rs324981 was lower than that of the controls. By studying the six main haplotypes (Table 3) which comprised 89.7% of all haplotypes in the samples, no association between SNP haplotypes and asthma could be identified. Nonetheless, haplotype H3 (CTACGA) was found to be closely related to tIgE level in the case-control survey (P=0.004).
| SNP ID/Allele | Nucleotide position NT_007819.18 | Type | Shandong Cases/Controls# (N=143/N =171) | Population | |||
|---|---|---|---|---|---|---|---|
| Finnish (N=499) | Canadian (N=402) | German (N=3113) | Chinese (N=232) | ||||
| rs323917 (C/G) | 34702031 | G | 0.09/0.07 | 0.05 | 0.06 | 0.06 | NA* |
| rs323920 (C/T) | 34707405 | T | 0.30/0.29 | NA | NA | NA | 0.27 |
| rs324377 (A/C) | 34716363 | A | 0.45/0.43 | 0.47 | 0.44 | 0.45 | NA |
| rs324396 (C/T) | 34750511 | C | 0.53/0.81 | 0.66 | 0.76 | 0.69 | 0.79 |
| rs324957 (A/G) | 34761760 | G | 0.51/0.48 | 0.47 | 0.47 | NA | 0.53 |
| rs324981 (A/T) | 34778501 | A | 0.31/0.51 | 0.47 | 0.47 | NA | 0.49 |
Table 2. Comparison of allelic distribution of SNPs from GPR154 in subjects from Shandong Province and other populations.
| Haplotypes | Frequency | Odds ratio (95% Confidence interval, P) | |
|---|---|---|---|
| Asthma | High total IgE levels | ||
| H1 CCATAA | 37.60% | 0.85 (0.63-1.12, 0.36) | 1.17 (0.48-2.52, 0.61) |
| H2 CCCCGA | 21.50% | 0.91 (0.74-1.30, 0.64) | 0.89 (0.62-1.34, 0.24) |
| H3 CTACGA | 11.20% | 1.05 (0.57-1.66, 0.47) | 1.53 (1.09-2.65, 0.004) |
| H4 CCCCAT | 10.90% | 1.14 (0.68-2.09, 0.85) | 0.96 (0.58-1.63, 0.79) |
| H5 CTCTAT | 8.50% | 1.22 (0.83-2.17, 0.52) | 0.77 (0.52-1.24, 0.38) |
Table 3. Relationship between haplotypes for GPR154 and asthma as well as 90th percentile of IgE levels in case-control study of Shandong subjects.
Subsequently, association between asthma, tIgE levels and single SNP genotypes were investigated by employing logistical regression. As a result (Table 4), rs324396 and rs324981 were related to asthma (P=0.003 and 0.002, respectively) and high total IgE levels (P=0.01 and 0.007, respectively). Notably, rs324981 was the solo nonsynonymous SNP which would cause a substitution, producing Asn instead of Ile. The changed sequence and resultant structure may affect the function of GPR154 gene which in turn, was associated with asthma.
| SNP ID | Genotype | Cases* (%) | Controls* (%) | Asthma Odds ratio (95% Confidence interval, P) | High total IgE levels (95% Confidence interval, P) |
|---|---|---|---|---|---|
| rs323917 | CC | 110 (76.9) | 134 (78.4) | 1 | 1 |
| CG | 29 (20.3) | 35 (20.5) | 0.43 (0.19-0.68, 0.26) | 1.14 (0.52-2.57, 0.64) | |
| GG | 1 (0.7) | 2 (1.2) | 0.78 (0.13-3.24, 0.67) | 0.35 (0.09-1.12, 0.27) | |
| CC | 85 (59.4) | 90 (52.6) | 1 | 1 | |
| rs323920 | CT | 14 (9.8) | 19 (11.1) | 0.97 (0.43-1.50, 0.85) | 0.49 (0.17-1.23, 0.11) |
| TT | 44 (30.8) | 61 (35.7) | 0.91 (0.46-1.83, 0.24) | 1.16 (0.62-1.81. 0.43) | |
| CC | 53 (37.1) | 65 (38.0) | 1 | 1 | |
| rs324377 | AC | 70 (49.0) | 79 (46.2) | 0.73 (0.45-1.26, 0.41) | 0.27 (0.08-0.84, 0.35) |
| AA | 18 (12.6) | 27 (15.8) | 1.04 (0.86-1.38, 0.92) | 0.92 (0.35-1.94, 0.31) | |
| CC | 59 (41.2) | 60 (35.1) | 1 | 1 | |
| rs324396 | CT | 55 (38.4) | 68 (39.7) | 1.02 (0.67-1.42, 0.86) | 0.73 (0.39-1.27, 0.19) |
| TT | 25 (17.4) | 42 (24.5) | 0.42 (0.15-0.70, 0.003) | 0.53 (0.31-0.92, 0.01) | |
| GG | 51 (35.7) | 64 (37.4) | 1 | 1 | |
| rs324957 | GA | 60 (41.9) | 68 (39.8) | 0.71 (0.38-1.47, 0.26) | 0.65 (0.42-1.38, 0.13) |
| AA | 30 (20.9) | 38 (22.2) | 2.04 (0.54-13.23, 0.44) | 0.86 (0.48-1.45, 0.24) | |
| TT | 49 (34.3) | 52 (30.4) | 1 | 1 | |
| rs324981 | TA | 63 (44.1) | 70 (40.9) | 0.87 (0.43-1.94, 0.62) | 0.57 (0.24-1.08, 0.27) |
| AA | 29 (20.3) | 48 (28.1) | 0.34 (0.26-0.81, 0.002) | 0.62 (0.34-0.95, 0.007) |
Table 4. The association between GPR154 gene polymorphisms and asthma as well as 90th percentile of high total IgE levels in the case-control analysis.
Discussion
Since the initial identification of GPR154 by positional cloning to be an asthma susceptibility gene, it has also been suggested various association with mental disorder, high IgE levels, airway hyperresponsiveness and inflammatory bowel disorder and NPS-induced calcium signaling pathways [21-23]. However, among the independent studies on distinct ethnic populations, some of the conclusions did not verify the relationship between the SNPs and the disease of interest [12,24]. For deciphering the genuine function of GPR154, duplicating such researches and studying different populations for more eloquent evidence are indispensable.
Phenotype evaluation is crucial for genetic experiments on sophisticated diseases including asthma. Typically, diagnosis of asthma is based upon classic methods which feature the recurrent episodes of wheezing, coughing and breathlessness as well. In our study, we used the combination of physician’s diagnosis and elevated airway responsiveness to obtain suitable and appropriate subjects.
We selected six SNPs, which is in line with two previous studies [10,16]. For the six major haplotypes presented, H3 (CTACGA) indicated its association with high levels of tIgE. However, no significant relationship between haplotypes and asthma was found. Although it is still widely debated about the efficiency and usefulness of single-SNP analysis compared with haplotype studies, we utilized both for analysis. For rs324981, the minor allele homozygotes AA lowered the risk for asthma (case-control OR: 0.34, P=0.002) and tIgE levels (OR: 0.62, P=0.007). rs324396 (TT) was protective for asthma (OR: 0.42, P=0.003) and tIgE levels (OR: 0.53, P=0.01). Since several SNPs were investigated in this study, we adopted Benjamini-Hochberg (BH) method to control the false discovery rate. After multiple comparisons using R platform, the above-mentioned P values were adjusted to 0.018, 0.06, 0.018 and 0.06, respectively.
As a common implementation for genetic researches on trying diseases, the case-control design aimed at the subpopulation in Shandong Province, China where this stable population in history and singular ethnic group provided unbiased foundation for the current research.
In all, our studied unambiguously indicates two SNPs rs324981 and rs324396 are associated with the occurrence of asthma, in a Chinese population. By doing so, we highlight again the important role of GPR154 involving in asthma and allergies. However, for studying the relationship between the two SNPs and high total IgE levels, further work with a larger sample size is needed.
Acknowledgements
None
Conflict of Interest
The authors declare no conflict of interest.
References
- Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev 2011; 242: 10-30.
- Olin JT, Wechsler ME. Asthma: pathogenesis and novel drugs for treatment. BMJ 2014; 349: g5517.
- Sharma S, Ghosh B. Promoter polymorphism in the MS4A2 gene and asthma in the Indian population. Int Arch Allergy Immunol 2009; 149: 208-218.
- Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES. Genome-wide search for asthma susceptibility loci in a founder population. The collaborative study on the genetics of asthma. Hum Mol Genet 1998; 7: 1393-1398.
- Weiss ST, Raby BA. Asthma genetics 2003. Hum Mol Genet 2004; 13: 83-89.
- Sharma M, Batra J, Mabalirajan U, Sharma S, Nagarkatti R, Aich J. A genetic variation in inositol polyphosphate 4 phosphatase A enhances susceptibility to asthma. Am J Resp Crit Care 2008; 177: 712-719.
- Laitinen T, Polvi A, Rydman P, Vendelin J, Pulkkinen V. Characterization of a common susceptibility locus for asthma-related traits. Science 2004; 304: 300-304.
- Laitinen T, Daly MJ, Rioux JD, Kauppi P, Laprise C. A susceptibility locus for asthma-related traits on chromosome 7 revealed by genome-wide scan in a founder population. Nat Genet 2001; 28: 87-91.
- Erdmann F, Kugler S, Blaesse P, Lange MD, Skryabin BV, Pape HC. Neuronal expression of the human neuropeptide S receptor NPSR1 identifies NPS-induced calcium signaling pathways. Plos One 2015; 10: 0117319.
- Vergara C, Jimenez S, Acevedo N, Martinez B, Mercado D, Gusmao L. Association of G-protein-coupled receptor 154 with asthma and total IgE in a population of the Caribbean coast of Colombia. J Br Soc Allergy Clin Immunol 2009; 39: 1558-1568.
- Kormann MSD, Carr D, Klopp N, Illig T, Leupold W, Fritzsch C. G-protein-coupled receptor polymorphisms are associated with asthma in a large German population. Am J Resp Crit Care 2005; 171: 1358-1362.
- Wu H, Romieu I, Sienra-Monge JJ, del Rio-Navarro BE, Burdett L, Yuenger J. Lack of association between genetic variation in G-protein-coupled receptor for asthma susceptibility and childhood asthma and atopy. Genes Immun 2008; 9: 224-230.
- Shin HD, Park KS, Park CS. Lack of association of GPRA (G protein-coupled receptor for asthma susceptibility) haplotypes with high serum IgE or asthma in a Korean population. J Allergy Clin Immun 2004; 114: 1226-1227.
- Soderhall C, Marenholz I, Nickel R, Grüber C, Kehrt R. Lack of association of the G protein-coupled receptor for asthma susceptibility gene with atopic dermatitis. J Allergy Clin Immunol 2005; 116: 220-221.
- Plaza V, Bellido-Casado J, Alonso-Coello P, Rodrigo G. Asthma clinical practice guidelines: advantages and pitfalls. Arch Bronconeumol 2009; 45: 25-29.
- Feng Y, Hong X, Wang L, Jiang S, Chen C, Wang B. G protein-coupled receptor 154 gene polymorphism is associated with airway hyperresponsiveness to methacholine in a Chinese population. J Allergy Clin Immunol 2006; 117: 612-617.
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing 2015.
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005; 21: 263-265.
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J. The structure of haplotype blocks in the human genome. Science 2002; 296: 2225-2229.
- Melen E, Bruce S, Doekes G, Kabesch M, Laitinen T. Haplotypes of G protein-coupled receptor 154 are associated with childhood allergy and asthma. Am J Respir Crit Care Med 2005; 171: 1089-1095.
- Pietras CO, Vendelin J, Anedda F, Bruce S, Adner M, Sundman L. The asthma candidate gene NPSR1 mediates isoform specific downstream signalling. BMC Pulm Med 2011; 11: 39.
- Hersh CP, Raby BA, Soto-Quiros ME, Murphy AJ, Avila L, Lasky-Su J. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Resp Crit Care Med 2007; 176: 849-857.
- Blakey JD, Sayers I, Ring SM, Strachan DP, Hall IP. Positionally cloned asthma susceptibility gene polymorphisms and disease risk in the British 1958 Birth Cohort. Thorax 2009; 64: 381-387.
- Veal CD, Reynolds NJ, Meggitt SJ, Allen MH, Lindgren CM, Kere J. Absence of association between asthma and high serum immunoglobulin E associated GPRA haplotypes and adult atopic dermatitis. J Investig Dermatol 2005; 125: 399-401.