Research Article - Virology Research Journal (2017) Virology Research Journal (Special Issue 1-2017)
Studies on the molecular variability in Indian isolates of Papaya ringspot virus
Shyam Singh1, Anjeet Jangre1, Pankaj Kumar1, Awasthi LP2*1S.K. College of Agriculture and Research Station, Kawardha, Chhattisgarh, India
2Amity Center for Research and Innovation(Amity University Uttar Pradesh),Varanasi-221007(U.P.),India
- Corresponding Author:
- Awasthi LP
Amity Center for Research and Innovation(Amity University Uttar Pradesh)
Varanasi-221007
India
Tel: 09415718904 (P)
Phone: 91-522 2668993 (R)
E-mail: lpawasthi06@yahoomail.com
E-mail: lpawasthi14@gmail.com
Accepted Date: January 17, 2017
Citation: Singh S, Jangre A, Kumar P, et al. Studies on the molecular variability in Indian isolates of Papaya ring spot virus. Virol Res J. 2017;1(1):10-16.
Abstract
Papaya ringspot virus (PRSV), Potyvirus is an important pathogen of papaya that causes severe losses in papaya production globally. The coat protein (CP) genes of four PRSV isolates originating from different locations in India, were cloned and sequenced. The band was observed at 840 bp in PRSV-FZD isolates of Uttar Pradesh. The isolates of PRSV have been characterized as papaya-infecting (PRSV-P). The PRSV-FZD sequence has made cluster with PRSV-Lucknow (AY458620). While sequences from Himanchal Pradesh (AY458617), Sikkim (DQ354072) clustered in another group with Jharkhand isolates (AY458619). Sequences from Karnataka (AY458618), Tamilnadu (DQ077175) and West Bengal (AY238885) have clustered in other group. The cluster analysis of the PRSV-FZD sequences and other Indian isolate has further done with PRSV sequences from different geographical regions. The results showed that PRSV-FZD had aligned in a cluster with other Indian sequences except sequences from Varanasi (AY238882), Maharashtra (AY238881), Madhya Pradesh (DQ650651) and Andhra Pradesh (AY903266) which clustered in separate groups. The sequences from Vietnam, China, Australia and Japan form a separate cluster. Brazil and USA sequences clustered in separate group along with the divergent Indian group.
Keywords
Poty virus, Pathogen, Chymopapain, Isolates, cluster
Introduction
Papaya (Carica papaya L.), a popular fruit tree of tropical and subtropical countries of the World, is considered as one of the rich source of vitamins A and C. In addition, it contains enzyme papain and chymopapain, both of which are widely used in the food and drug [1]. It is consumed as fresh ripen fruit as well as vegetable and also in the preparation of value added products. However, the total fruit production is declining as this crop is badly affected by wide range of biotic factors such as fungi, bacteria, viruses, and nematodes. Among them, viruses are the major limiting factor in the cultivation of papaya in many countries including India, especially northern India. A large number of viruses have been reported from time to time on papaya which belong to cucumo-, gemini-, ilar-, poty-, rhabdo-, tobra- and tospo- virus groups [2]. Among them Papaya ring spot virus (PRSV), a member of the genus Potyvirus of family Potyviridae [3], is a major and ubiquitous limiting factor for papaya production worldwide. It causes severe yield losses (20%) in papaya production. PRSV also infects many cucurbits and other crop plants [3]. Particles of PRSV are flexuous rods measuring 760 nm to 800 nm × 2 nm [4]. It consists of positive sense single stranded RNA with 9000 to 10,326 nucleotides in length excluding the poly ‘A’ tail [5] encapsidated by 30 kD to 36 kD coat protein. According to the host range specificity, PRSV is classified into two biotypes: (i) PRSV-W, formerly water melon mosaic virus 1, which naturally infects many Cucurbitaceous crops but is unable to infect papaya; and (ii) PRSV-P, which naturally infects papaya (Carica papaya) and can be transmitted experimentally to cucurbits as well. In addition, a strain of PRSV (PRSV-T) isolated from squash in Gaudeloupe does not infect papaya and is reported to be biologically and serologically different from PRSV-P and PRSV-W. Most isolates of these two pathotypes are serologically indistinguishable when tested by coat protein (CP) and cylindrical inclusion protein (CIP) specific antisera. The PRSV isolates have not been well characterized at the molecular level. In order to determine the sequence diversity within PRSV isolates from different geographical locations of India, coat protein (CP) nucleotide and amino acid sequences of PRSV isolates were analyzed. This report is focused on the initial molecular characterization of the CP gene of four PRSV strains and sequence comparison to other reported PRSV sequence at both the nucleotide and amino acid levels.
Material and Methods
Isolation and purification of causal viruses
Isolation and purification of plant viruses was performed to separate virus particles from plant constituents. Highly purified preparations are essential for chemical analysis of the viruses but less pure virus will often suffice to produce an antiserum. Techniques for purification aim at the separation of virus from host constituents without affecting the structure and infectivity of the virus to be purified. PRSV was purified by the protocol of Gonsalves and Ishii [6] with little modifications. One hundred grams fresh leaves of papaya plants infected with PRSV were collected, washed with water, blotted dry, cut into small pieces and frozen overnight at -20°C. The frozen leaves were homogenized in 0.5 M potassium phosphate buffer, pH 7.5 supplemented with 0.01 M ethylene diamine tetra acetate (EDTA) and 0.1% sodium sulphate (PE buffer). This slurry was stirred continuously for 2 hrs at 4°C and then extracted through a triple layer of cheese cloth. The filtrate was clarified with a mixture of 5% chloroform + 5% carbon tetrachloride (1:1 v/v) for 15 minutes. The clear solution thus obtained was first subjected to low speed centrifugation (5,000 g) for 15 min in a Servall RC-5B centrifuge followed by super speed centrifugation using SE-12 or SA-600 fixed angle rotors. Servall OTD-65 B ultracentrifuge was used for high speed centrifugation in A-841 fixed angle rotor for final pelleting of virus particles at 4°C and TST 41.14 swing out rotor was used for the density gradient run. The resulting emulsion of previous step was centrifuged at 5,000 g for 10 minutes at 4°C. The aqueous phase was carefully pipetted out from centrifuge tubes. Eight gram of polyethylene glycol (PEG) MW-600 was added to the aqueous phase and the suspension was centrifuged at 10,000 g for 10 minutes. The pellets obtained were resuspended in 20 ml PE buffer (0.1 M KPO4 buffer containing 0.01 M EDTA), pH 7.0. The solution was centrifuged at 10,000 g for 10 minutes and virus was reprecipitated by adding PEG and NaCl to final concentration of 5% and 0.3 M, respectively. The solution was centrifuged at 10,000 g for 10 minutes. Supernatant was loaded on to 535 (w/w) CS2SO4 pad prepared in 0.5 M PE buffer and centrifuged at 10,000 for 10 min. The virus containing zone was collected and diluted with two volumes of PE buffer. The pellet thus obtained was suspended in 0.1 M PE buffer in 1/5 of total volume and kept at 4°C for overnight. The pellet was then dissolved and centrifuged at low speed (5,000 g) for 10 minutes. The supernatant was again pelletized at 35,000 g for 2 hrs and the pellet was resuspended in 0.05 M PE buffer. The final virus pellet was suspended in 0.01 M potassium phosphate buffer, pH 8.0.
Virus identity
Papaya seedlings, inoculated with infected papaya leaf tissue were observed daily for the occurrence of viral symptoms. The plants showing viral disease symptoms were assayed for the production of the viral coat protein by standard DAC-ELISA procedures [7].
Reverse transcript-polymerase chain reaction (RT-PCR)
The virus isolate infecting papaya, identified as PRSV was further characterized by reverse transcript-polymerase chain reaction (RT-PCR) and the nucleotide and amino acid sequence was compared with available similar sequence in gene data bank. The coat region from PRSV isolates collected from Faizabad was amplified in two steps using two sets of primers. Overlapping fragments representing partial CP regions was amplified using HRP 52 (5'TCCAARAATGAAGCTGTGGATGT3') and RKJ 3 (5'GTTGCGCATA CCCAGGAGAG3') primers [8]. The amplification conditions in both the steps included one cycle of reverse transcription for 45 min at 42°C, 40 cycles of denaturation for 30s at 94°C, annealing for 2 min at 45°C and extension for 60 min at 72°C. The cDNA synthesized by the above process was checked on 1% agarose gel electrophoresis. PCR amplified DNA gel was purified using a PCR purification kit (Qiagen) and then cloned in pGEM-T easy vector (Promega). Two clones of each isolate were sequenced in both directions and the sequencing was carried out at the Department of Biochemistry, South Campus, University of Delhi, Delhi, India. The sequences of the gene were assembled using ‘Edit Seq program’ of the Laser gene 6.0 (DNASTAR Inc., Madison, WI, U.S.A.). Sequence identity among all different PRSV isolates as determined, and deduced amino acids sequences were assembled into multiple sequence alignment using Clustal W program. Phylogenetic tree was constructed using the DNA star, Mega Align or Mega 3.1 program. Alignment was done by Clustal method by using the parameters: Gap penalty-10, Gap length penalty-10, Pair wise alignment was done by the parameters: Ktuple-2, Gap penalty-5, Window-4, Diagonal saved-4. The sequences were compared by taking last 840 nucleotides of already submitted sequences. Nucleotide sequence were compared with the CP sequences from India and other Asian, North American, South American.
Sequence analysis and comparison
Both nucleotide as well as amino acid sequences of four PRSV CP-coding regions were compared with already available CP sequences of PRSV isolates from other countries isolates using the Martinez/ Needleman-Wunsch and the Clustal algorithms from the Meg Align (version 3.03) program (DNASTAR). Multiple sequence analyses were made using the CLUSTAL W program [9].
Results
Virus identity
The virus identity was confirmed by pathogenicity tests in an insect-proof glasshouse. The virus inoculum, prepared by homogenizing infected papaya leaf tissue in 0.01 mol/L potassium phosphate buffer (pH 7.0) containing sodium sulphite (1%, w/v), was applied by gentle rubbing onto the leaves of healthy papaya plants. Differential plants showing viral symptoms were assayed for the production of the viral coat protein by standard double-antibody sandwich enzyme-linked immunosorbent assays (DAS-ELISA) procedures [10]. Four PRSV isolates reacted with the commercial PRSV antibody, three isolates (Faizabad, Etawah, Lucknow) had a stronger reaction, whereas one (Varanasi) isolate reacted weakly. Inoculation experiments indicated that these isolates produced visible symptoms in papaya. All papaya seedlings inoculated with PRSV inocula, showed typical viral symptoms (mosaic and leaf distortion) one month after inoculation. These isolates of PRSV were characterized as papaya-infecting (PRSV-P).
Reverse transcription-polymerase chain reaction (RTPCR)
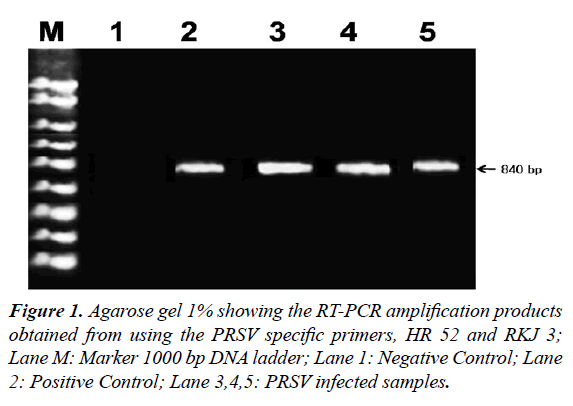
Amplification of PRSV-FZD isolate viral RNA using CP gene specific primers by RT-PCR resulted in the production of an amplicon of about 840 bp fragments (Figure 1).
Sequencing of coat protein gene
The sequence data revealed that the PRSV-FZD gene (Table 1) had an ORF of 855 bp, which could potentially encode a protein of 285 amino acids with an approximate molecular weight of 36 kDa. The results of the BLAST (www.ncbi.nih.gob/BLAST) search performed to identify sequence homology, clearly demonstrated that the sequence matched with the reported PRSV coat protein gene sequences from different geographical locations.
| 1 | TCGAAGGCTG | AAGCTGCGGA | TGCGGGCTCA | AATGACAAGC | TCAAAGAGAG | AGAACAGAAA |
| 61 | GAGAAGGAGA | AGAAGAAAGA | GGAAGAAAAA | GATGAAGCTA | GTGGTGTAGA | TGATGTGTCA |
| 121 | TCGACTAGCA | CGAAAACTGG | AGAGAGAGAT | AGAGATGTTA | ACGCTGGAAC | TAGTGGAACT |
| 181 | TTTACCAGTCC | CAAGGATAAA | ATCGTTTACT | GACAAGATGG | CACAGCAAAT | AATTAAGGGA |
| 241 | AAAACTGTCC | TTAATTTGGA | TCATCTTCTT | CAGTATAACT | AGGGGGTGAG | TGACATCTCA |
| 301 | AACACTCGTG | CTACCCAATC | TCAGTTTGAG | AGGTGGTACG | TGATGGTATG | GGATGATTAT |
| 361 | GGTCTTAACG | ACAGTGAAAT | GCAAGTGATG | TTAAATGGTT | TGGATGGCGA | GTGTATTGAA |
| 421 | AATGGTACAT | CTCCAGACAT | ATCTGGTGTC | TGGGTAATGA | CATTTACTGA | CAATCAAGTC |
| 481 | GACTATCCGA | TCAAGCCTTT | GATTGAACAT | GCAACTCCTT | ATGCTACTGA | GATCATGGCT |
| 541 | CACTTCAGTA | ACGCGGCAGA | AGCATACATT | GCAAAGAGAA | TCGCTAGATA | GAGATACATG |
| 601 | CCGCGGTATG | GAATCAAGAG | AAATTTGACT | GACATTAGCC | AAGCTCACAT | CGCTTTCGAT |
| 661 | TTCTATGAGG | TGAATTCGAA | AACACCTGAT | AGGGCTCGCG | AAGCTCACAT | GCAGCTGAAA |
| 721 | GCTGCGGCAT | TGCGAAACAC | TAATCGCAGA | ATGTTTGGCA | TGGACGGCAG | TGTCAGTAAC |
| 781 | AAGGAAGAGA | ACACGGAGAG | ACACACAGTA | GAAGATGTCA | ATAGAGACAT | GCGCTCTCTC |
| 841 | CTGGGTATGC | GCAAC |
Table 1: Partial nucleotide of coat protein region of PRSV-FZD isolate.
Phylogenetic study
For the study of phylogenetic relation of the PRSV-FZD to other PRSV isolates reported from India and worldwide, their nucleotide sequences were compared using ‘DNA STAR’ programme and phylogenetic tree was prepared (Table 2). In this study, last 840 bp of selected sequences were aligned using Clustal method with residue weighted table. They share percent similarity from 81.4% to 97.7%. The maximum percent similarity of PRSV-FZD was observed with the Lucknow isolate (AY458620)-97.7% followed by Haryana isolate (DQ088670) with 91.7% similarity. Also, the maximum divergences of 14.5% and 14.2% were found with Karnataka (AY458618) and Tamilnadu (DQ077175) isolates, respectively.
| Percent Similarity | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| percent Divergence | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |||
| 1 | 87.1 | 86.8 | 85.5 | 88.2 | 89.9 | 91.7 | 88.1 | 95.5 | 86.4 | 92.4 | 86.0 | 88.2 | 1 | AY238881-MAR-IND.SEQ | ||
| 2 | 14.2 | 88.0 | 91.2 | 87.4 | 85.3 | 84.3 | 89.0 | 85.1 | 89.3 | 85.4 | 87.0 | 85.1 | 2 | AY238881-MAR-IND.SEQ | ||
| 3 | 12.7 | 14.0 | 83.6 | 94.3 | 93.6 | 83.9 | 94.4 | 84.5 | 84.2 | 84.4 | 93.2 | 91.2 | 3 | AY458617-HP-IND-SEQ | ||
| 4 | 11.7 | 6.9 | 15.0 | 83.7 | 85.1 | 84.6 | 85.4 | 85.8 | 94.4 | 85.5 | 82.4 | 83.6 | 4 | AY458618-KR-IND-SEQ | ||
| 5 | 11.6 | 14.0 | 5.9 | 14.7 | 93.3 | 85.7 | 96.1 | 86.1 | 83.9 | 85.7 | 93.6 | 91.2 | 5 | AY458619-JHK-IND-SEQ | ||
| 6 | 9.1 | 13.7 | 6.4 | 12.9 | 6.8 | 86.7 | 94.2 | 87.1 | 85.2 | 85.1 | 91.9 | 97.7 | 6 | AY458620-LUK-UP-IND-SEQ | ||
| 7 | 5.7 | 14.0 | 12.7 | 12.2 | 12.0 | 10.2 | 85.8 | 91.5 | 85.1 | 91.5 | 83.7 | 84.3 | 7 | AY903266-KP-IND-SEQ | ||
| 8 | 11.7 | 13.1 | 5.8 | 13.3 | 4.1 | 5.9 | 11.2 | 86.7 | 84.9 | 86.2 | 93.1 | 91.7 | 8 | DQ088570-HAR-IND-SEQ | ||
| 9 | 4.7 | 15.1 | 15.5 | 12.7 | 14.3 | 12.1 | 5.7 | 13.2 | 85.0 | 91.5 | 85.1 | 85.5 | 9 | DQ650651-MP-IND-SEQ | ||
| 10 | 12.4 | 9.5 | 14.7 | 4.6 | 14.9 | 13.5 | 11.9 | 14.1 | 13.2 | 85.4 | 81.4 | 83.6 | 10 | DQ077175-TN-IND-SEQ | ||
| 11 | 6.1 | 14.8 | 14.1 | 12.6 | 13.4 | 12.0 | 6.9 | 12.5 | 6.9 | 13.7 | 84.4 | 84.4 | 11 | DQ354071-OG-IND-SEQ | ||
| 12 | 13.8 | 14.9 | 7.1 | 16.7 | 6.7 | 8.5 | 13.3 | 7.4 | 14.7 | 16.0 | 14.6 | 89.9 | 12 | DQ354072-SIKKIM-IND-SEQ | ||
| 13 | 10.0 | 14.5 | 7.7 | 13.5 | 7.8 | 1.1 | 11.3 | 7.1 | 13.1 | 14.2 | 12.7 | 9.2 | 13 | PRSV-FZD-IND-SEQ | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | ||||
Table 2: Percent similarity and divergence of PRSV-FZD isolate with other Indian isolates.
The similarity with respect to isolates from other geographical regions from world varied (Table 3) from 82.5% to 85.4%. The maximum similarity was found with Japan isolates (D50591) 85.4% followed by China (DQ449536) 85.1%, Brazil (AY094985) 83.5%, USA (NC-001785) 83.5% and Vietnam (AF506889) 83.2%. Pakistan isolate (AB127935) with 69.5% similarity and Australia (AF506902) having 82.5% have least similarity with PRSV-FZD isolates. Evidently the Pakistan isolate have maximum divergence (28.8%) from PRSV-FZD isolate followed by Australia (15.6%).
| Percent Divergence | Percent Similarity | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||||
| 1 | 70.8 | 71.3 | 70.6 | 70.6 | 71.5 | 70.6 | 72.6 | 72.3 | 69.8 | 70.2 | 70.6 | 71.8 | 69.6 | 69.2 | 71.9 | 69 | 71.2 | 70 | 69.5 | 1 | AB127935-PAKISTAN.S | ||
| 2 | 26.7 | 90.8 | 88.5 | 93.2 | 94.4 | 88.5 | 87.7 | 87.1 | 84.8 | 84 | 85 | 85.1 | 84.6 | 88.1 | 86.3 | 87.5 | 85.8 | 84.5 | 83.2 | 2 | AF-506889-VIEPNAM.SE | ||
| 3 | 28.2 | 9.5 | 87.6 | 93.2 | 91.5 | 87.6 | 87.1 | 86.5 | 83.9 | 83.9 | 83.2 | 84.1 | 84.2 | 86.2 | 85.4 | 86.7 | 83.9 | 82.5 | 82.6 | 3 | AF-506902-AUSTRALIA | ||
| 4 | 28.5 | 11.7 | 12.1 | 89.5 | 89.9 | 100 | 92.1 | 91.4 | 84.4 | 82.7 | 85 | 83.3 | 84.5 | 91.3 | 845.4 | 91.7 | 85.7 | 83.3 | 83.6 | 4 | AY-09485-BRAZIL.SEQ | ||
| 5 | 27.7 | 6.6 | 6.9 | 10.4 | 94.5 | 89.5 | 88.5 | 87.6 | 86.4 | 86.3 | 85.8 | 85.4 | 85.8 | 88.5 | 88 | 89.3 | 87.1 | 86 | 85.4 | 5 | D50591-JAPAN.SEQ | ||
| 6 | 26.2 | 5.8 | 8.6 | 9.6 | 5.1 | 89.9 | 89.2 | 88.5 | 86.8 | 85.7 | 86.3 | 86.5 | 86 | 88.5 | 88 | 89.4 | 87.3 | 85.5 | 85.1 | 6 | DQ-449535-CHINA.SEQ | ||
| 7 | 28.5 | 11.7 | 12.1 | 0 | 10.4 | 9.6 | 92.1 | 91.4 | 84.4 | 82.7 | 85 | 83.3 | 84.5 | 91.3 | 84.4 | 91.7 | 85.5 | 83.3 | 83.5 | 7 | NO-001785-USA.SEQ | ||
| 8 | 27.4 | 12 | 12.4 | 7.6 | 11.4 | 10.6 | 7.6 | 99.2 | 87.1 | 86.8 | 86.5 | 88.2 | 89.9 | 91.7 | 88.1 | 95.5 | 86.4 | 86 | 88.2 | 8 | AY-238881-MAR-IND.SEQ | ||
| 9 | 27.9 | 12.6 | 12.9 | 7.8 | 12.1 | 11.3 | 7.8 | 0.4 | 86.4 | 86.3 | 86.4 | 87.7 | 89.4 | 91.7 | 87.4 | 94.6 | 86.6 | 85.2 | 87.5 | 9 | AY238882-VAR-UP-IND | ||
| 10 | 31 | 14.8 | 15.4 | 16.3 | 13.7 | 12.8 | 16.3 | 14.1 | 14.7 | 88 | 91.2 | 87.4 | 88.3 | 84.3 | 89 | 86.1 | 89.3 | 87 | 86.1 | 10 | AY238885-WB-IND-SEQ | ||
| 11 | 28.9 | 15.4 | 15.3 | 17.1 | 13.8 | 13.7 | 17.1 | 13.1 | 13.4 | 13.8 | 83.6 | 94.3 | 93.6 | 83.9 | 94.4 | 84.5 | 84.2 | 93.2 | 91.2 | 11 | AY458517-HP-IND-SEQ | ||
| 12 | 29.3 | 14.3 | 15.7 | 13.9 | 13.7 | 12.2 | 13.9 | 12.2 | 12.5 | 6.9 | 14.9 | 83.7 | 85.1 | 84.6 | 85.4 | 85.8 | 94.4 | 82.4 | 83.6 | 12 | AY458618-KAR-IND.SEQ | ||
| 13 | 27.2 | 14.2 | 15.4 | 16.7 | 13.8 | 13 | 16.7 | 11.8 | 12.1 | 13.8 | 5.9 | 14.5 | 93.3 | 85.7 | 96.1 | 86.1 | 83.9 | 93.6 | 91.2 | 13 | AY458619-JHK-IND.SEQ | ||
| 14 | 28.7 | 14.1 | 14.8 | 14.3 | 12.6 | 12 | 14.3 | 9.5 | 9.8 | 13.5 | 6.4 | 12.8 | 6.8 | 86.7 | 94.2 | 87.1 | 85.2 | 91.9 | 97.7 | 14 | AY458620-LUK-UP-IND | ||
| 15 | 29.4 | 10.3 | 12 | 7.5 | 10 | 9.9 | 7.5 | 5.6 | 6.1 | 13.5 | 12.7 | 12 | 12.1 | 10.2 | 85.8 | 91.5 | 85.1 | 83.7 | 84.3 | 15 | AY903266-AP-IND-SEQ | ||
| 16 | 27.4 | 12.5 | 13.9 | 15.4 | 12 | 11.3 | 15.4 | 12.1 | 12.7 | 12.9 | 5.8 | 13.3 | 4.1 | 5.9 | 11.2 | 86.7 | 84.9 | 93.1 | 91.7 | 16 | BQ088670-HAR-IND.SEQ | ||
| 17 | 30.6 | 12.1 | 12.8 | 8.2 | 10.8 | 10.1 | 8.2 | 4.7 | 5.1 | 15.3 | 15.8 | 13.1 | 14.5 | 12.5 | 5.9 | 13.5 | 86 | 85.1 | 85.5 | 17 | DQ650651-MP-IND.SEQ | ||
| 18 | 27.8 | 13.8 | 15.3 | 14.1 | 13.2 | 11.6 | 14.1 | 12.9 | 13.1 | 9.5 | 14.6 | 4.6 | 14.8 | 13.4 | 11.7 | 14.1 | 13.6 | 81.4 | 43.6 | 18 | DQ077175-TN-IND.SEQ | ||
| 19 | 28.6 | 15.1 | 16.6 | 16.5 | 13.8 | 13.4 | 16.5 | 14.2 | 14.8 | 14.7 | 7.1 | 16.6 | 6.7 | 8.5 | 13.3 | 7.4 | 15 | 15.9 | 89.9 | 19 | DQ354072-SIKKIM-IND-SEQ | ||
| 20 | 28.8 | 14.9 | 15.6 | 14.8 | 13.6 | 12.5 | 14.8 | 10.4 | 10.5 | 14.6 | 7.7 | 13.5 | 7.8 | 1.1 | 11.1 | 7.1 | 13.4 | 14.1 | 9.2 | 20 | PRSV-FZD-IND.SEQ | ||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||||
Table 3: Percent similarity and divergence of PRSV-FZD isolate with other isolates of the world.
The PRSV-FZD sequence made cluster with PRSV-Lucknow (AY458620). While sequences from Himanchal Pradesh (AY458617), Sikkim (DQ354072) clustered in another group with Jharkhand isolates (AY458619). Sequences from Karnataka (AY458618), Tamil Nadu (DQ077175) and West Bengal (AY238885) were clustered in other group.
The cluster analysis of the PRSV-FZD sequence and other Indian isolate was further done with PRSV sequences from different geographical regions. Results showed that PRSVFZD had aligned in a cluster with other Indian sequences except sequences from Varanasi (AY238882), Maharashtra (AY238881), Madhya Pradesh (DQ650651) and Andhra Pradesh (AY903266) which clustered in separate groups. The sequences from Vietnam, China, Australia and Japan form a separate cluster. Brazil and USA sequences clustered in separate group along with the divergent Indian group.
Discussion
The translated coat protein sequence indicated the presence of the conserved DAG sequence potentially associated with aphid transmissibility of many Potyviruses. CP gene sequence of 131 isolated (P=93; W=35; Uncharacterized=3) from different parts of the world including India, are now available in the GeneBank (Table 2). Comparison of the available CP gene sequences revealed that PRSV isolate vary in CP gene length from 840 to 870 nucleotides, encoding 280 to 290 amino acids. Thirty-one CP gene sequences have been submitted to Gene Bank in India. Considerable heterogeneity in CP gene length is also observed in Indian isolates, which varied from 840 to 867 nucleotides, encoding proteins of 280 (KA2 isolate) to 289 (Indore isolate) amino acids amino acids. The differences in CPs length were confined to the EK (Glutamic acid and Lysine) regions in amino terminal regions [11].
Jain et al. characterized PRSV isolates type P and type W from India (Pune) [12]. A 1.7 kb of the 3-terminal region comprising a part of the nuclear inclusion b(Nib) gene, the complete capsid protein and untranslated region (UTR) of both P and W isolates were cloned and sequenced. Comparative sequence analysis showed that the 3 UTRs in isolates P and W were 209 nucleotides in length excluding the ploy (A) tail, and shared 96% identity. The CP genes of two isolates were also similar to 87% nucleotide identity and 93% amino acid identity. They observed that the amino acid differences between CP genes were mostly confined to the amino terminus of the gene. The DAG triplet associate with aphid transmissibility was present in the CP of isolates W but it was replaced with DAD in the P isolate. The partial sequenced NIb genes were also 90% identical but isolate W contained an additional amino acid (threonine) just upstream of the protease cleavage site (Q/S) between NIb and CP. Compared to known PRSV isolates, the Indian isolates exhibited greater sequence divergences in their CP genes.
Kunklikar and Byadgi characterized PRSV isolates type from India (Dharwad) [13]. They reported that nucleotide sequence of coat protein gene of Dharwad isolate had an A + T content of 58.74% and G +C content of 41.26%. The amino acid composition data showed different percentage of amino acids. However, the coat protein had particularly low amount of methionine (0.45%) and high amount of serine (9.09%). The sense strand molecular weight as DNA was 222.23 kDa and antisense strand molecular weight as DNA was 219.54 kDa. The weight of DNA duplex was 441.78 kDa. The sense strand molecular weight as RNA was 231.1 kDa and antisense strand molecular weight as RNA was 227.66 kDa. The weight of RNA duplex was 458.76 kD. The nucleotide frequencies observed was adenine-237 (33.1%), guanine-120 (16.8%) cytosine-175 (24.50%) and thymine 183 (25.6%). The AT density was more at N-terminal region as compared to C-terminal. Studies on CpG islands revealed that there was high density of CpG islands towards C-terminal starting from core region of the CP gene. There results indicated that the amino region of Dharwad code protein gene had more nucleotide variation as compared to the core region of different PRSV isolates. The N-terminal region had more EK (Glutamine and lysine) repeats starting from the beginning of the sequence. The similarity with respect to EK repeats was also observed in isolates of Indian origin. The Dharwad isolate was found to cluster with other isolates of Indian origin and Australian isolates, sowing a similarity of 97.60% and 95.6%, respectively. The similarity of Dharwad isolates with respect to isolates from other geographical region was America-92.3%, Maxico-93.9%, Hawaii-95.10%, India-93.9%, India-93.91%, Brazil-94.4%, Japan-91.3%, Taiwan-90.6%, Indonesia-90.0%, Thailand-90.9%, Vietnam-87.9% and India-88.2%.
Papaya ring spot virus genome was sequenced completely from four overlapping cDNA clones and by direct sequencing of viral RNA [14]. The genomic RNA 10326 nucleotides in length excluding the poly (A) tract contains one large open reading frame that starts are nucleotide position 86 to 88 and ends that position 10118 to 10120 encoding a polyprotein of 3344 amino acids. The highly conserved sequenced AAAUAA AANANCUCAACACAACAVA at the 5' end of the RNA of PRSV and those of the other five reported Potyviruses showed 80% similarity suggesting that this region may play a common important role for Potyvirus replication. Two cleavage sites of polyprotein were determined by amino acid sequencing of the N termini of helper component (HC-Pro, amorphous inclusion) and cylindrical inclusions (CI) proteins. Other cleavage sites were predicted by analogy with other Potyviruses. The genetic organization of PRSV in similar to that of other Potyviruses except that the first protein processed from the N terminus of the polyprotein (NT) has an MW of 62 K, 18 K to 34 to layer than those of other Potyviruses. The cleavage site was liberating N terminus of the HC-Pro protein was found at the same location downstream from the consensus sequence FI (V) VRG as that reported for tobacco vein mottling Potyvirus. The NT protein of Potyvirus appears to be the NIb protein, the putative polymerase for the replication of the polyviral RNA. The genetic organization of PRSV RNA is tentatively proposed to be VPa-5' leader 63 K NT52 K, HC PRO-46 K-72, CI-6 K-48 K, Mia-59 K, NIb-35 K coat protein-3', non-coding region-poly (A) tract.
Coat protein derived resistance has been effectively used to confer resistance to a wide range of viruses. Its success depends on relatedness of transgenes with challenge virus strain [15,16]. It is therefore essential to compare the nucleotide amino acid sequences of coat protein (CP) gene to determine the level of variability among PRSV strains. The coat protein gene lies in the carboxyl terminal end of the Potyvirus polyprotein. Comparison of coat protein sequences can provide evidence for variability in strains or isolates which may have significant effect on the level of stability of transgenic resistance.
Wang and Yeh reported nucleotide sequence comparisons of the 3' terminal region of severe, mild, and non-papaya infecting strains of PRSV [15]. The 3' terminal 2561 nucleotide residues of the severe HA strain of PRSV was determined. Comparison with the published sequence of the mild strain PRSV HA 5-1 showed that the two strains shared a 99.4% identity in the 3' terminal 22-35 residues. They differed in 10 residues at the Nib gene, resulting in five amino acid changes, and in two residues in the code protein gene, resulting in five amino acid changes. The 3' un-translated regions were identical but HA contain 2 more nucleotides (AG) at the 3' extreme. Comparisons with the published non-papaya infecting type W strain PRSV-W revealed a 97.9% similarity in the 3' terminal 2235 residues. Strains PRSV-W and HA differed in 40 nucleotides in the coding region, resulting in four amino acid changes in the NIb gene and 6 in the CP gene and also differed in 7 nucleotides in the 3' untranslated region [8,11].
The nucleotide sequence comparison revealed that the N-terminal of the PRSV code protein is highly variable as previously described for other Potyviruses [8,11,16-18]. The core and C-terminal region were most conserved. The variability in N-terminal region was most evident in first 38 amino acids that contain a stretch of EK (Glutamine and Lysine) repeats staring at the third amino acid after DAG aphid transmission motif [11,18]. Jain et al. reported that the amino terminal region of PRSV like other Potyviruses were extremely variable in the first 50 amino acids [11].
Silav Rossels et al. reported that the comparison of the coat protein sequence of three Mexican isolates of PRSV with other geographical isolates showed a close relationship to American and Australian isolates. The CP gene of the PRSV was cloned and sequenced in three Mexican isolates and these sequences were compared to 11 isolates from other parts of the world. They were found to have higher similarity to isolates from Australia and United State than to Asian isolates. A region of about 100 nucleotide residues neighbouring the putative aphid transmission triplet of the coat protein contained repeats of an EK (Glutamic acid-Lysine) modify in all the sequences. It was suggested that this region could have a bearing on the genetic relationship and geographical distribution of the various isolates.
Comparative sequence analysis of PRSV isolates from different countries at amino acid level revealed that there was low substantial correlation of CP sequence diversity with geographical origin of isolates [11]. The isolates from India and Bangladesh shared 89% to 100% sequence identity, while other Asian isolates and the isolates from Australia and Americas (BZ, MX and US) were more closely related to each other (93% to 97% identity). Amongst Indian isolates, maximum variation at amino acid levels was shown by CG, DL, DI-W, JK, KA2, UP-LK and WB isolated (up to 11%) followed by AP, HP, KA1 and KA3 isolates (upto 7%). On the other hand, PUM, PU-S and UP-V isolates showed least variation (up to 3%) cluster dendogran based on nucleotide and deduced amino acid sequences also revealed that clustering of the PRSV isolates did not correlate with their geographical origin [11]. PRSV isolates were grouped into major clusters. The Asian isolates other than the Indian and Bangladesh isolates (CH, JAP, PHP, TW, TH and VN) formed one cluster. The remaining isolate formed second cluster, which were further differentiated into three sub-clusters. Those from central (CG), eastern (JK, WB) and northern (DL, UP, -LK) India and BD isolate from Bangladesh formed one cluster, while the isolates from northern (UP-V) and western (PU-M, PU-S) India, Australia (AUS), Americas (BZ, MX, US) formed second cluster and the third was a cluster of isolates from southern India (HP, DL-W). The Indian isolates grouped into two separate groups without any geographical correlation thus appear to be a single mixed population. Similarly, the isolates from America (USA and Brazil) clustered in one group. While, the Asian isolates (Vietnam, China, Australia, and Japan) clustered into separate group, the isolates from Pakistan did not aligned to any of the group.
The CP sequences of 25 PRSV isolates originating from different countries were compared. The conserved regions in the CP of Potyvirus such as WCIEN and UMKAAA were present in all the isolates. A stretch of KE (lysine and glutamic acid) repeats (KE region) was also observed in the amino terminal region for all the PRSV sequence analyzed. Further, the DAG triplet, attributed to the aphid transmissibility of the virus, was also conserved in the CP of 11 of the 14 Indian isolates [19].
Considerable heterogeneity in the CP length was observed and the CP-coding region in the Indian isolates varied from 840- 867 nucleotides, encoding proteins of 280 (Karnataka isolate) to 289 (Indore) amino acids. Size differences presented from the differences in the number of KE repeats in the amino terminal region. Like other Potyviruses [8], the amino terminal region was extremely variable in the first 50 amino acids and the most amino acids substations were identical in this region.
As sown earlier [8,19,20], present study clearly revealed substantial sequence variation (upto 28.8%) within PRSV population worldwide. The most divergent isolates are the Asian isolates (upto 28.8%). Amongst the Asian isolates, the Indian isolates are as different from themselves (14.5%) as they are from, other Asian (12.5% to 28.8%), Australian (15.6%) and Americas (12.5% to 14.8%). The present study in accordance with earlier reports [8,11,19,20], clearly showed that the Asian isolates have higher divergence, which is further increased in Indian isolates. Thus, the Asia in general and India in particular can be regarded as area of origin of PRSV [8,11,19,20], due to presence of sub-population. The higher divergence exhibited by the Indian isolates could be attributed to wide range of cropping systems.
This study will have significant role in devising PRSV management strategy in the country through the CP gene derived transgenic resistance or mild strained cross protection. Since, selection of trans gene is crucial for development of virus resistance transgenic papaya in the country.
References
- Frazier WC, Westhoff DC. Food Microbiology. 4th ed. Tata McGraw-Hill Publication Company Limited: New Delhi; 2007.
- Purcifull DE, Hiebert E. Serological distinction of Watermelon mosaic virus isolates. Phytopathology. 1979;69:112-6.
- Tripathi S, Suzuki JY, Ferreira SA, et al. Papaya ringspot virus-P: Characteristics, pathogenicity, sequencing variability and control. Mol Plant Pathol. 2008;9(3):269-80.
- Yeh SD, Gonsalves D. Purification and immunological analysis of cylindrical inclusion protein induced by Papaya ringspot virus and Watermelon mosaic virus-1. Phytopathol. 1984;74:1273-78.
- Wang CH, Yeh SD. Divergence, and conservation of the genomic RNA’s of Taiwan and Hawaii strains of Papaya ringspot Potyvirus. Arch Virol. 1997;142:271-85.
- Gonsalves D, Ishii M (1980) Purification and serology of Papaya ringspot virus. Phytopathol. 1980;70:1028-32.
- Lakshminarayana Reddy CN. Studies on Papaya ring spot virus disease on papaya (Carica papaya L.). M.Sc. (Ag.) Thesis, University of Agricultural Sciences, Bangalore. 2000.
- Jain RK, Sharma J, Verma A. Present status of Papaya ringspot virus population profile in India. Annu Rev Plant Pathol. 2004a;3:1-15.
- Thomson JD, Higgins DG, Gibson TJ. CLUSTUR W: Improving the sensitivity of progressive multiples sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673-80.
- Converse RH, Martin RR. ELISA methods for plant viruses. In: Serological methods for detection and identification of viral and bacterial plantpathogens. R. Hampton, E. Ball, and S. De Boer, editors. American Phytopathological Society,St. Paul, MN. 1990;179-96.
- Jain RK, Sharma J, Sivakumar AS, et al. Variability in the coat protein gene of Papaya ring spot virus isolates from multiple locations of India. Ach Virol. 2004b;149:2425-442.
- Jain RK, Pappu HR, Pappu SS, et al. Molecular characterization of Papaya ringspot virus isolates (P and W) from India. Ann Appl Bio. 1998;132:413-25.
- Kunklikar S, Byadgi AS. Molecular characterization, cloning of coat protein gene epidemiology and management of Papaya ringspot virus. In “Record development in the diagnosis and management of plant disease for meeting global challenges” held at University of Agricultural Sciences, Dharwad, 18-20 December. 2003.
- Yeh SD, Jan FJ, Chiange CH, et al. Complete nucleotide sequence and genetic organization of Papaya ringspot virus RNA. J Gen Virol. 1992;73:2531-41.
- Wang CH, Yeh SD. Nucleotide sequence comparison of the 3’-termianl regions of severe, mild and non-papaya infecting strains of Papaya ringspot virus. Arch Virol. 1992;127:133-40.
- Bateson M, Henderson J, Chaleeprom W, et al. Papaya ringspotvirus Potyvirus: Isolate variability and origin of PRSV type P. (Australia). J Gen Virol. 1994;75:3547-53.
- Wang CH, Bau HJ, Yeh SD. Comparison of the nuclear inclusion b protein and coat protein gene of five Papaya ringspot virus strains distinct in geographic origin and pathogenicity. Phytopathol. 1994;84(10):1205-10.
- Hema MV, Prasad DT. Comparison of the coat protein of a south Indian strain PRSV with other strains from different geographical locations. J Plant Pathol. 2004;86(1):35-2.
- Byadgi AS. Papaya ringspot virus. In: characterization, diagnosis, and management of plant viruses Vol. 1. Industrial Crops.Rso GP, Khurana SMP, Lenardan S, editors. Studium Press: USA; 2006.
- Bateson MF, Rosemarei EL, Revill P, et al. On the evaluation and molecular epidemiology of the PotyvirusPapaya ringspot virus. J Gen Virol. 2002;83:2575-85.