Commentary - Journal of RNA and Genomics (2017) Volume 13, Issue 1
Rethinking Delivery: Viral Vectors and RNA Interference
Jens Kurreck*
Department of Applied Biochemistry, Institute of Biotechnology, Berlin University of Technology, Germany
- *Corresponding Author:
- Jens Kurreck
E-mail: jens.kurreck@tu-berlin.de
Tel: +49 30 314 27582
Fax: +49 30 314 27502
Received date: 07 March 2017; Revised date: 16 March 2017; Accepted date: 20 March 2017; Published date: 25 March 2017
© Copyright The Author(s). First Published by Allied Academies. This is an open access article, published under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0). This license permits non-commercial use, distribution and reproduction of the article, provided the original work is appropriately acknowledged with correct citation details
Introduction
Efficient delivery of nucleic acids to their intended target cells remains the major complication holding back the widespread adoption of RNA interference (RNAi) therapies. In recent years, substantial progress has been made in the development of delivery systems for small interfering RNAs (siRNAs), including lipid formulations, nanoparticles and functional groups covalently coupled to siRNAs. So far the vast majority of clinical trials involving RNAi therapies have relied on the use of such chemically generated systems. Some organs, however, remain difficult to target with currently available delivery agents. Viral vectors are a promising alternative to deliver short hairpin RNA (shRNA) expression cassettes. Many corporate researchers are reluctant to consider viral vectors as a means of delivery due to the poor reputation of gene therapy, as a consequence of adverse events that occurred in early studies. More recently developed vector systems have excellent safety profiles as a result of systematic improvements in this key area. The time has come to re-consider viral vectors in order to overcome the complications and allow efficient, specific and well-tolerated delivery in RNAi applications.
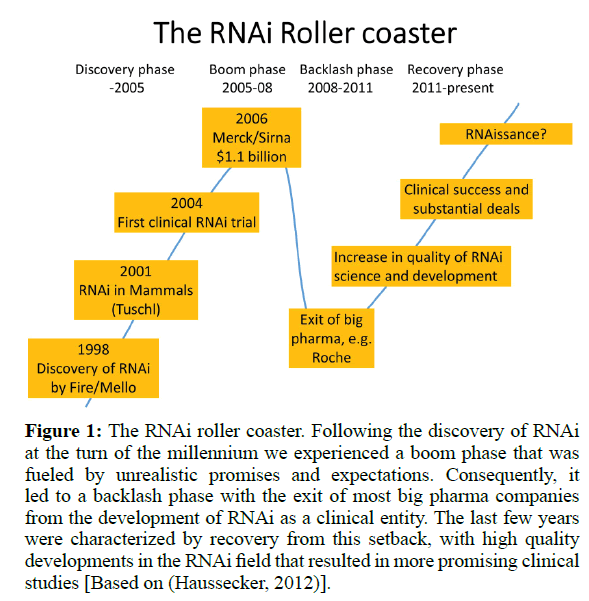
Following the discovery that RNAi can be employed in mammalian cells for the specific silencing of a gene-of-interest (Elbashir et al, 2001; Kurreck, 2009) the field experienced an incredible boom. It took only three years until the first clinical trial based on RNAi treatment was initiated. What followed can be denoted as the RNAi roller coaster illustrated in (Figure 1). The enthusiasm culminated in the acquisition of the biotech company SiRNA Therapeutics by Merck for $1.1 billion in 2006. At that time, much unsubstantiated hope was placed in the technology. It was inevitable that many of these expectations could not be fulfilled and the RNAi field experienced a backlash phase with spectacular exits of major pharmaceutical companies (“big pharma”). Although disappointing to many, in the long run this was a healthy development. Since then the quality of RNAi science and development increased substantially, resulting in clinical success which, in turn, laid the foundation for some substantial new financial deals. Although still annotated with a big question mark, Nature Biotechnology already coined the term RNAissance for the development in 2014 (Editorial, 2014).
Figure 1: The RNAi roller coaster. Following the discovery of RNAi at the turn of the millennium we experienced a boom phase that was fueled by unrealistic promises and expectations. Consequently, it led to a backlash phase with the exit of most big pharma companies from the development of RNAi as a clinical entity. The last few years were characterized by recovery from this setback, with high quality developments in the RNAi field that resulted in more promising clinical studies [Based on (Haussecker, 2012)].
What are the reasons for the ups and downs of RNAi technology? One issue is the limited specificity of siRNAs. However, this seems to be more of a problem for scientific studies than for medical applications of the technology. While it is important to ascribe the effects of depletion of a gene-ofinterest to its specific knockdown in a scientific publication, we know that no drug will ever be fully specific for its target and “dirty” compounds have proven to be even more effective in (cancer) therapy than highly specific ones. Thus, the main challenge in RNAi applications still remains what Nobel prize winner Phillip Sharp already proclaimed in 2003 and what has become the Holy Trinity of RNAi: “The major hurdle right now is delivery, delivery, delivery” (Check, 2003). Consequently, much effort has been put into the development of improved methods for the delivery of siRNAs. We now have a selection of strategies for siRNA transfer, including lipid formulation and covalent coupling of lipophilic substances that support crossing the membrane barrier. While many delivery systems are non-specific, some allow specific delivery to the targeted tissue by coupling of antibodies, aptamers or receptor ligands to the siRNA or nanoparticle used for delivery (Kurreck, 2009; Haussecker, 2014). Conjugation of N-Acetylgalactosamine (GalNAc) to siRNAs is considered one of the most promising covalent modifications (Zimmermann et al, 2017).
Some of these approaches have proven to be efficient; however, it is no surprise that the oligonucleotides often end up in the liver (particularly those coupled to GalNAc), the primary organ for detoxification of exogenous substances. This explains why a large share of current clinical RNAi trials addresses hepatic diseases. Admittedly, even though hepatitis C is now sometimes considered a “cured disease” with the approval of new drugs such as Sovaldi® (Sofosbuvir) (Nakamura et al, 2016), there are more than enough liver diseases for which new treatment options are still impatiently awaited. Still, we will have to develop strategies to target other organs and tumors elsewhere in the body.
This is where gene therapy comes into play, another field that has experienced ups and downs in its development. Initially, it had the charm that if genes can cause disease, then they will also be suited to treating maladies. In the late 80s and early 90s, people tended to make irresistible promises to heal previously devastating diseases. This led to some unpleasant incidents, when half-baked approaches were used in humans and ethical borders were crossed to fulfill investors’ expectations and for the hoped for honor of being the first to reach a new level. This development culminated in the well-known fatal incident when a young man died after having been treated with an inappropriately high dose of an adenoviral vector (Lehrman, 1999). In other cases, technical issues resulted in the development of leukemia in children that were treated with retroviral vectors which integrated in the vicinity of oncogenic genes (Hacein-Bey- Abina et al, 2003). While these events were unbearably sad, they also led to a shakeup in the field. With improved methods for gene transfer, success returned so that the first approval of a gene therapy approach (in the Western world) finally took place in 2012 (Yla-Herttuala, 2012): Glybera is based on an Adeno-Associated Virus (AAV) vector to treat lipoprotein lipase deficiency. In 2016, the first ex vivo gene therapy was approved by European authorities. The active component of Strimvelis is a lentiviral vector used to treat patients suffering from severe combined immunodeficiency due to a defect in the Adenosine Deaminase Gene (ADA-SCID). The production of viral vectors is expensive: The cost of treatment with Glybera exceeds $1 million per patient. However, this number must be compared to the price of conventional treatment. For example, enzyme replacement therapy of ADA-SCID costs $4.25 million per patient for 10 years. In addition, the cost of gene therapeutic drugs can be expected to drop as they become more common. The same happened for the large scale synthesis of oligonucleotides, which once seemed prohibitively expensive and can now be produced at a reasonable price.
In 2002, several groups independently developed systems to express short double-stranded RNA molecules intracellular instead of transfecting chemically synthesized siRNAs (Shi, 2003). These shRNAs are processed by the cellular machinery into siRNAs, which then knockdown the target gene. This strategy not only allows prolonged silencing, as the shRNAs are continuously produced, it also opens up the possibility of using viral vectors for the delivery of double-stranded RNA molecules. This inspired the leading AAV experts Dirk Grimm and Mark Kay to entitle their 2007 review “RNAi and Gene Therapy: A Mutual Attraction” (Grimm and Kay, 2007). The authors saw an enormous potential in combining the efficacy of RNAi-mediated silencing with methods for efficient and tissue-specific gene transfer by viral vectors that had been under development for two decades. Since then, many groups used the vector approach for various scientific RNAi applications, including transduction of hard-to-transfect cells (e.g. primary cells), the generation of stable cell lines and transgenic knockdown animals, and genome-wide screens.
This extensive use in research applications contrasts with the rare clinical use of vectors inducing RNAi-silencing. In a recent review, Martinez et al. listed 26 siRNA-based programs in clinical development (Martinez et al, 2015). In contrast, only one vector-based trial for the treatment of HCV infection was mentioned. According to the Benitec homepage, this program is going to be terminated as a “result of the increasingly competitive landscape” (benitec.com as of February 6th, 2017). Two further small trials with vectors delivering shRNAs against HIV and HBV, respectively (Haussecker, 2012), are also not being developed further.
What is the reason for the discrepancy between the extensive successful use of the vector-shRNA approach in research applications and the prevalence of siRNA usage in clinical trials? It is the author’s experience from many conferences that particularly representatives of larger biotech companies and major pharmaceutical companies, who could sponsor clinical trials, are adamantly opposed to the use of viral vectors. This strategy has a poor reputation based on the negative experiences made 15 to 20 years ago. The significant improvements in viral vectors that have been made since then have largely been ignored. Chemists put much effort into the development of new delivery agents that are efficient, non-toxic and preferably tissue-specific. Nature spent eons to optimize its strategies for the highly efficient and, in many cases, specific delivery of transgenes. We decided to call the resulting nanoparticles “viruses”. There is no question that viruses can cause severe disease and many adverse effects caused by the use of viral vectors have been seen. However, newer generations of viral vectors have greatly improved safety, e.g. lentiviral vectors with self-inactivating LTRs (Kohn and Candotti, 2009).
Among the most promising delivery vehicles for transgenes are AAV vectors (Daya and Berns 2008). To date, no known pathology is associated with (natural) AAVs, which is the first advantage of this vector type compared to others. Their capability to integrate into the genome of their host is (virtually) eliminated by removing the required genes from the viral genome, which abolishes the risk of genomic insertion. AAVs induce a mild immune reaction, making fatal adverse effects, such as those observed for adenoviral vectors, unlikely. One restriction of AAV vectors is their limited packaging capacity of 4.7 kb. Self-complementary viral vectors, which are the most common form used today due to their rapid onset of transgene expression and high expression level, can only use half this length for their primary sequence. However, while this limitation is disadvantageous for the expression of large genes, it is not a restriction for RNAi-approaches, as shRNA expression cassettes are usually less than 500 nucleotides in length, i.e. AAV vectors are even large enough to transfer multiple shRNA expression cassettes in parallel, if needed. Another valuable feature of AAV vectors is their directable tissue tropism. More than 100 AAV serotypes are currently known, 10 of which are widely used in biomedical research (Lisowski et al, 2015). While the genome usually contains elements such as the inverted terminal repeats (ITRs) of the standard serotype 2, the DNA can be packaged into capsids of any of the other serotypes. This procedure, called pseudotyping, can be used to direct the vector to the intended target tissue. We have, for example, shown that an AAV vector consisting of a capsid of serotype 9 (denoted as AAV2.9) can efficiently transduce cardiomyocytes following i.v. injection into mice and silence the expression of an endogenous gene (Suckau et al, 2009) or inhibit a heart pathogenic virus (Stein et al, 2015). In contrast, all attempts to deliver chemically synthesized siRNAs formulated with a lipid delivery agent to cardiomyocytes failed. It should be noted that none of the viral vectors is absolutely specific for a single tissue. The above mentioned serotype 9 not only transduces cardiomyocytes, but also gets into the liver and skeletal muscle. However, additional features, such as tissue-specific promoters or microRNAtarget sites, can be used to increase the specificity of transgene expression from an AAV vector (Geisler and Fechner, 2016).
Thus, it seems time to reassess the prejudices against viral vectors resulting from negative experiences in the distant past and to consider them as an option for efficient and safe delivery of genetic material. We now know that there is no magic RNAi bullet, but instead we will have to find optimized solutions for each indication with regard to efficiency, tissue specificity, time to onset of the silencing and its duration (acute vs. chronic diseases). To fulfill all these requirements, we need as many options as possible and viral vectors should be regarded as an opportunity for clinical RNAi applications without bias in order to add to the current repertoire of chemical delivery systems.
Regarding the current drug development landscape, it is reasonable to assume that it will be small and medium-sized biotech enterprises that will drive the development of viral vectors for delivery purposes. As they will make progress in early phases of clinical testing, big pharma will enter the field and acquire promising candidates. It will also be important to watch the developments in conventional gene therapy to transfer progress there into the RNAi field as rapidly as possible. If several further approvals take place, the potential of the field will become apparent to even the most reluctant, leading to an increase in interest and investment which will accelerate progress even more.
Acknowledgements
The author wants to thank Erik Wade for careful proof-reading and sharing visionary thoughts that contributed to the article.
References
- Elbashir SM, Harborth J, Lendeckel W, et al. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494-498
- Kurreck J. 2009. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed Engl, 48, 1378-1398
- Editorial. 2014. Is this really the RNAissance? Nat Biotechnol, 32, 201
- Check E. 2003. Gene regulation: RNA to the rescue? Nature, 425, 10-12
- Haussecker D. 2014. Current issues of RNAi therapeutics delivery and development. J Control Release, 195, 49-54
- Zimmermann TS, Karsten V, Chan A, et al. 2017. Clinical Proof of Concept for a Novel Hepatocyte-Targeting GalNAc-siRNA Conjugate. Mol Ther, 25, 71-78
- Nakamura M, Kanda T, Haga Y, et al. 2016. Sofosbuvir treatment and hepatitis C virus infection. World J Hepatol, 8, 183-190
- Lehrman S. 1999. Virus treatment questioned after gene therapy death. Nature, 401, 517-518
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. 2003. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science, 302, 415-419
- Yla-Herttuala S. 2012. Endgame: Glybera finally recommended for approval as the first gene therapy drug in the European Union. Mol Ther, 20, 1831-1832
- Shi Y. 2003. Mammalian RNAi for the masses. Trends Genet, 19, 9-12
- Grimm D and Kay MA. 2007. RNAi and Gene Therapy: A Mutual Attraction. Hematology Am Soc Hematol Educ Program, 2007, 473-481
- Martinez T, Jimenez AI and Paneda C. 2015. Short-interference RNAs: becoming medicines. EXCLI J, 14, 714-746
- Haussecker D. 2012. The Business of RNAi Therapeutics in 2012. Mol Ther Nucleic Acids, 1, e8
- Kohn DB and Candotti F. 2009. Gene therapy fulfilling its promise. N Engl J Med, 360, 518-521
- Daya S and Berns KI. 2008. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev, 21, 583-593
- Lisowski L, Tay SS, Alexander IE. 2015. Adeno-associated virus serotypes for gene therapeutics. Curr Opin Pharmacol, 24, 59-67
- Suckau L, Fechner H, Chemaly E, et al. 2009. Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy. Circulation, 119, 1241-1252
- Stein EA, Pinkert S, Becher PM, et al. 2015. Combination of RNA interference and virus receptor trap exerts additive antiviral activity in coxsackievirus B3-induced myocarditis in mice. J Infect Dis, 211, 613-622
- Geisler A, Fechner H. 2016. MicroRNA-regulated viral vectors for gene therapy. World J Exp Med, 6, 37-54.