Research Article - Biomedical Research (2017) Volume 28, Issue 19
Regulation roles of lncRNA SPRY4-IT1 on the growth and migration of osteosarcoma
Boming Zhao1,2, Kebin Liu2, Xiaobin He2, Jiyuan Yang2, Yang Yi2 and Lin Cai1*
1Department of Orthopedic Surgery, Zhongnan Hospital Affiliated to Medical College of Wuhan University, Wuhan, Hubei, PR China
2Department of Orthopaedic Surgery, the No. 1 People's Hospital of Jingzhou, Jingzhou, Hubei, PR China
- *Corresponding Author:
- Lin Cai
Department of Orthopedic Surgery
Zhongnan Hospital Affiliated to Medical College of Wuhan University, PR China
Accepted date: August 22, 2017
Abstract
Objective: LncRNA SPRY4-IT1 usually plays an oncogenic role in various cancers. However, the function of SPRY4-IT1 in Osteosarcoma (OS) still hasn’t been reported. Our study aimed to investigate the regulation roles of SPRY4-IT1 in the OS.
Methods: OS tissue and the adjacent normal tissue were collected from 175 patients with OS. SPRY4- IT1 in OS cells was silenced by siRNA interference. The expression of SPRY4-IT1, Cyclin D1, Matrix Metalloproteinase-2 (MMP-2), Matrix Metalloproteinase-9 (MMP-9) and WNT/β-catenin signaling pathway-related factors-Wnt3 and Wnt5b in OS tissues and cells were detected by qRT-PCR and/or western blot. Effect of SPRY4-IT1 knockdown on growth, colony formation, migration and invasion of OS cells were detected by MTT assay, colony formation assay and cell migration and invasion assay, respectively. Correlation between expression of SPRY4-IT1 and expression of Cyclin D1, MMP-2 and MMP-9 was analysed by Pearson correlation analysis.
Results: Level of SPRY4-IT1 RNA in OS tissue was significantly higher than that in normal tissue. SPRY4-IT1 knockdown significantly reduced the viability, colony formation ability, and invasion and migration ability of OS cells. SPRY4-IT1 knockdown significantly decreased the expression of Cyclin D1, MMP-2, MMP-9 and WNT/β-catenin signaling pathway-related factors Wnt3 and Wnt5b at both mRNA and protein levels. In addition, expression of SPRY4-IT1 was positively correlated with the expression of Cyclin D1, MMP2 and MMP9.
Conclusion: SPRY4-IT1 plays an important role in the progression of OS possibly by regulating the expression of Cyclin D1, MMP-2, MMP-9 and WNT/β-catenin signaling pathway-related genes.
Keywords
SPRY4-IT, Osteosarcoma, Cyclin D1, MMP-2, MMP-9, WNT/β-catenin signaling pathway
Introduction
As the most common primary malignant tumor in bone, OS mainly affects adolescents and young adults [1]. About 10 persons out of a million people are suffering from OS, and 20 to 50% patients are in their pediatric age [2]. Although OS only accounts for 5-6% of all the tumors in children, the high metastasis rate makes it one of the most common causes of cancer-related death [3]. With the advances in the treatment of OS [4], 5 y survival rate has been increased from about 20% (1970s) to 65-70% [5]. However, this 5 y survival rate of patients with OS has not been further increased during last two decades, and almost half of the patients will eventually die of OS [6]. The onset and development of cancer is a complex process requiring the involvement of various factors, such as environmental and cellular factors [7]. It has been widely accepted that environmental factors, such as radiation, air pollutions an so on, can only affect the development of cancer by regulating the expression of cancer-related gene and/or regulating signaling transduction involved in this process [8-10]. So, the identification of molecular and cellular targets that involved in the development of OS will definitely provide new insights into the development of more efficient treatment of OS.
Long non-coding RNAs, also known as lncRNA, is a group of non-protein coding RNAs with a length longer than 200 nucleotides, which is obviously longer than that of microRNAs (miRNAs), short interfering RNAs (siRNAs) and other short RNAs [11]. Previous studies have shown that lncRNAs play pivotal roles in the development of various human diseases including heart disease, liver diseases and cancer [12-14]. SPRY4-IT1 is a long non-coding RNA that is transcribed from an intron of the SPRY4 gene. It has been reported that expression of lncRNA SPRY4-IT1 is usually upregulated in cancer tissues and the increased expression level of SPRY4- IT1 can promote tumorigenesis [15,16]. Studies also showed that SPRY4-IT1 could regulate tumor cell apoptosis and invasion [17]. Those previous studies suggest that SPRY4-IT1 may also play important roles in the development of OS.
In our study, the regulation roles of lncRNA SPRY4-IT1 in the growth and migration of OS were explored. We found that the expression level of SPRY4-IT1 RNA in OS tissue was significantly higher than that in normal tissue. SPRY4-IT1 knockdown by siRNA interference significantly reduced the viability, colony formation ability, and invasion and migration ability of OS cells. SPRY4-IT1knockdown also significantly decreased the expression of cell cycle regulator Cyclin D1, cell migration regulator MMP-2 and MMP-9, and WNT/β-catenin signaling pathway-related factors Wnt3 and Wnt5b at both mRNA and protein levels. In addition, expression of SPRY4- IT1 was found to be positively correlated with the expression of cyclin D1, MMP2 and MMP9. So, we believe that SPRY4- IT1 plays important roles in the development of OS possibly by regulating the expression of Cyclin D1, MMP-2, MMP-9 and WNT/β-catenin signaling pathway-related genes.
Materials and Methods
OS tissue collection
OS tissue and the adjacent normal tissue were collected from 175 patients with OS who received biopsy in Zhongnan Hospital Affiliated to Medical College of Wuhan University from January 2010 to October 2012. All patients received no treatment before admission. Tissues collected from patients were histologically and pathologically diagnosed by two experienced pathologists and confirmed that content of tumor tissue in each sample was higher than 70%. RNA was extracted from three sites of each tissue to destroy tumor heterogeneity. The median length of follow-up of these patients was 40 months.
Cell culture
Human osteosarcoma cell line MG63 and normal human bone cell line were purchased from The Cell Resource Center of the Institute of Basic Medical Sciences (IBMS) of The Chinese Academy of Medical Science. Cells were cultured in DMEM medium containing 10% FBS, 100 mg/ml of penicillin G and 100 U/ml of streptomycin (Invitrogen).
SiRNA transfection
SiRNA targeting SPRY4-IT1 (siRNASPRY4-IT1) and scrambled negative control siRNA were purchased from Shanghai Genepharma Pharmaceutical Technology Co., Ltd. Sequence of siRNASPRY4-IT1 was GCTTTCTGATTCCTGACTATTAAAGGCCTAGA. Cells were transfected with 50 nmol siRNASPRY4-ITl or scrambled negative control siRNA using lipofectamine 2000 transfection reagent (11668-019) (Invitrogen, Carlsbad, USA) according to the manufacturer's instructions. 48 h after transfection, transfected cells were harvested and transfection was checked by qRT-PCR.
MTT cytotoxicity test
Transfected MG63 cells were inoculated into 96-well plates with 200 μl per well and 10 μl of MTT (5 mg/ml in PBS) was added into each well. After incubation for 4 h, cell culture was terminated and supernatant was discarded. Cell suspension was centrifuged and the supernatant was removed again. Then 150 μl of Dimethyl Sulfoxide (DMSO) was added into each well, followed by oscillation for 10 min. OD value at 490 nm was measured by enzyme-linked immunosorbent assay analyzer.
RNA extraction and qRT-PCR
Trizol reagent (1 ml) (15596-026) (Invitrogen, Carlsbad, CA) was used to extract total RNA from tissue (100 mg) or harvested cells (2 × 106 cells) according to the manufacturer's instructions. Concentration and purity of RNA were determined by NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, USA). 1 μg RNA was used to synthesize cDNA through reverse transcription using PrimeScript™ One Step RT-PCR Kit (RR055A) (TaKaRa, Dalian, China) on 7300 RT-PCR system (Applied Biosystems, Carlsbad, USA). Following primers were used in PCR reactions: 5’-ATCCGAAGCGCAGACACAATTCA-3’ (sense) and 5’-CCTCGATGTAGTCTATGTCATAGGA-3’ (anti-sense) for SPRY4-IT; 5’- AGACTCGCTGATGATCCATGC-3’ (sense) and 5’-AGGTGACCACAGTGTTCTG-3’ (anti-sense) for GAPDH. PCR was performed as follows: 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 52°C for 30 s and 72°C for 1 min, then 10 min at 72°C.
Colony formation assay
Cell suspension (5 ml, containing 95% single cells) was seeded into a Petri dish after dilution of multiple proportions according to cell proliferation capacity. Cells were cultured at 37°C with 5% CO2 for 2-3 w. Culture medium was replaced with fresh medium based on changes in pH value of the medium. Finally, Giemsa (P0021) staining was used to observe cell colonies.
Cell migration and invasion assay
In order to measure cell migration ability, 5 × 104 transfected cells were seeded into the upper chamber (BD Biosciences, San Jose, USA). To measure cell invasion, 5 × 104 transfected cells were inoculated into the upper chamber covered with matrigel (356234) (Millipore, Billerica, USA). RPMI-1640 containing 20% FCS was used to fill the lower chamber. After incubation for 24 h, the membrane was stained with 0.5% crystal violet for 20 min. After removal of non-invasive cells, the invaded cells on membrane were counted under a microscope (Olympus, Tokyo, Japan).
Western blot
Total protein was extracted using Radioimmunoassay (RIPA) lysis buffer (P0013K, Beyotime). Protein lysates were separated by 10% SDS-polyacrylamide gel and transferred to a Polyvinylidene Difluoride (PVDF) membrane (Bio-Rad, Hercules, USA). After blocking, primary antibodies of cyclin D1 (#2978, Boston, USA), MMP2 (#13132, Boston, USA), MMP9 (#13667, Boston, USA) and GAPDH (#2118, Boston, USA) were incubated with the corresponding membrane overnight at 4°C. Then corresponding secondary antibody was added and incubated at 37°C for 1 h. Signal was detected with Enhanced Chemiluminescence (ECL) (WBKLS0050) and the grayscale intensity of each band was measured using bioimaging system (Millipore, Billerica, USA). This experiment was repeated three times.
Statistical analysis
SPSS19.0 software (SPSS, Inc., Chicago, Illinois, USA) was used for the data analysis. Normal distribution data were expressed by (͞x ± s), and comparisons between two groups were performed by t test. Non-normal distribution data were processed using non-parametric Mann-Whitney U test. p<0.05 was considered to be statistically significant.
Results
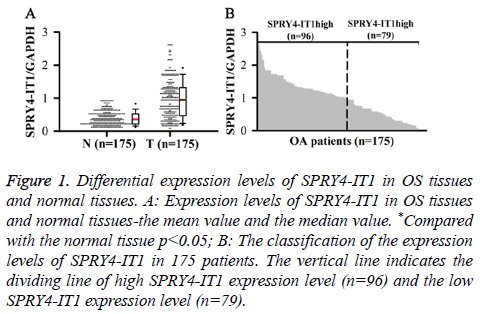
Differential expression levels of SPRY4-IT1 in OS tissues and normal tissues
SPRY4-ITl expression in OS Tissue (T) and Normal tissue (N) was detected by qRT-PCR. Relative expression levels of SPRY4-ITl were normalized to endogenous control gene (GAPDH). Our data showed that level of SPRY4-IT1 RNA in OS tissue was significantly higher than that in normal tissue (p<0.05) (Figure 1A). In addition, 175 patients were divided into high expression group and low expression group by a boundary value of the expression level of SPRY4-IT1/ GAPDH=1.014. We found that SPRY4-ITl was highly expressed in 96 out of 175 patients (Figure 1B).
Figure 1: Differential expression levels of SPRY4-IT1 in OS tissues and normal tissues. A: Expression levels of SPRY4-IT1 in OS tissues and normal tissues-the mean value and the median value. *Compared with the normal tissue p<0.05; B: The classification of the expression levels of SPRY4-IT1 in 175 patients. The vertical line indicates the dividing line of high SPRY4-IT1 expression level (n=96) and the low SPRY4-IT1 expression level (n=79).
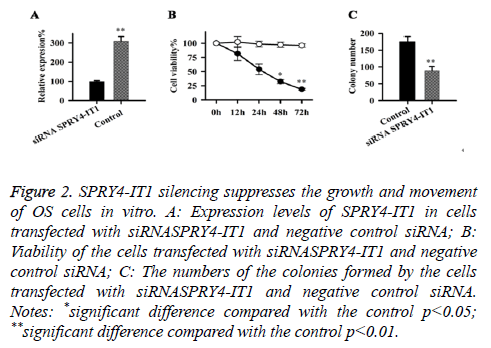
Effect of SPRY4-IT1 on the growth of OS cells
Expression of SPRY4-IT1 in cells transfected with siRNASPRY4-IT1 and negative control siRNA were detected by qRT-PCR, our data has shown that the expression level of SPRY4-IT1 in cells with siRNASPRY4-IT1 transfection was significantly lower than that in the cells with negative control siRNA (Figure 2A), indicating that SPRY4-IT1 was silenced by siRNASPRY4-IT1. Cell viability was measured by MTT assay. As shown in Figure 2B, the viability of the cells with siRNASPRY4-IT1 transfection was significantly lower than that of the cells transfected with negative control siRNA at all four time points but significant differences were only found at 48 h and 72 h (p<0.05, p<0.01). In addition, colony formation assay showed that the number of the colonies formed by cells with siRNASPRY4-IT1 transfection was significantly smaller than that formed by the cells transfected with negative control siRNA (p<0.01) (Figure 2C).
Figure 2: SPRY4-IT1 silencing suppresses the growth and movement of OS cells in vitro. A: Expression levels of SPRY4-IT1 in cells transfected with siRNASPRY4-IT1 and negative control siRNA; B: Viability of the cells transfected with siRNASPRY4-IT1 and negative control siRNA; C: The numbers of the colonies formed by the cells transfected with siRNASPRY4-IT1 and negative control siRNA. Notes: *significant difference compared with the control p<0.05; **significant difference compared with the control p<0.01.
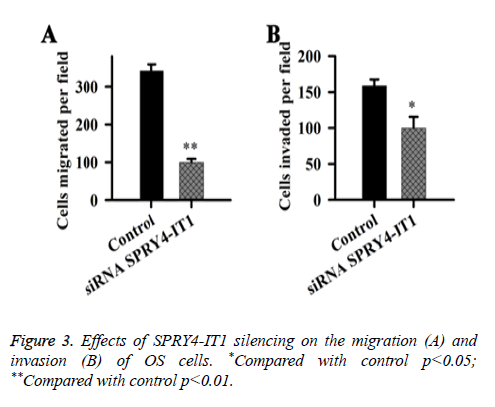
Effects of SPRY4-IT1 on the migration and invasion of OS cells
Effects of SPRY4-IT1 on the migration and invasion of OS cells were detected by cell migration and invasion assay. Data showed that the migration ability of the cells transfected with siRNASPRY4-IT1 was significantly lower than that of the cells transfected with negative control siRNA transfection (p<0.01) (Figure 3). In addition, the cell invasion ability of the cells transfected with siRNASPRY4-IT1 was also significantly lower than that of the cells transfected with negative control siRNA transfection (p<0.05).
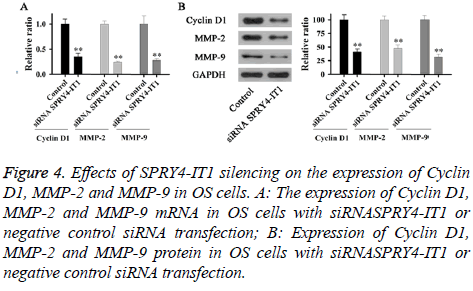
Effects of SPRY4-IT1 on the expression of Cyclin D1, MMP-2 and MMP-9 in OS cells
Expression of Cyclin D1, MMP-2 and MMP-9 mRNA and protein in OS cells with siRNASPRY4-IT1 or negative control siRNA transfection were detected by qRT-PCR and Western blot, respectively. As shown in Figure 4A, expression levels of Cyclin D1, MMP-2 and MMP-9 mRNA in cells with siRNASPRY4-IT1 transfection were significantly lower than those of the control cells (p<0.01). In addition, expression levels of Cyclin D1, MMP-2 and MMP-9 protein were also significantly lower than those of the control cells (Figure 4B).
Figure 4: Effects of SPRY4-IT1 silencing on the expression of Cyclin D1, MMP-2 and MMP-9 in OS cells. A: The expression of Cyclin D1, MMP-2 and MMP-9 mRNA in OS cells with siRNASPRY4-IT1 or negative control siRNA transfection; B: Expression of Cyclin D1, MMP-2 and MMP-9 protein in OS cells with siRNASPRY4-IT1 or negative control siRNA transfection.
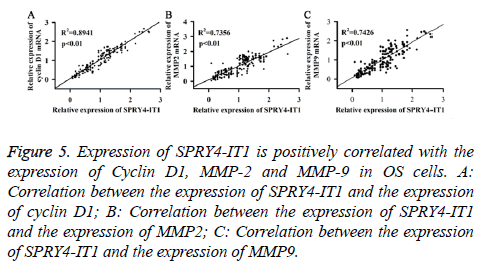
The correlation between the expression of SPRY4-IT1 and the expression of Cyclin D1, MMP-2 and MMP-9 in OS cells
Expression of SPRY4-IT1, Cyclin D1, MMP2 and MMP9 mRNA of the OS tissues from 175 patients was detected by qRT-PCR. Pearson correlation analysis was used to analyse the correlation between the expression of SPRY4-IT1 and the expression of cyclin D1, MMP2 and MMP9. As shown in Figure 5, expression of SPRY4-IT1 was positively correlated with the expression of cyclin D1, MMP2 and MMP9.
Figure 5: Expression of SPRY4-IT1 is positively correlated with the expression of Cyclin D1, MMP-2 and MMP-9 in OS cells. A: Correlation between the expression of SPRY4-IT1 and the expression of cyclin D1; B: Correlation between the expression of SPRY4-IT1 and the expression of MMP2; C: Correlation between the expression of SPRY4-IT1 and the expression of MMP9.
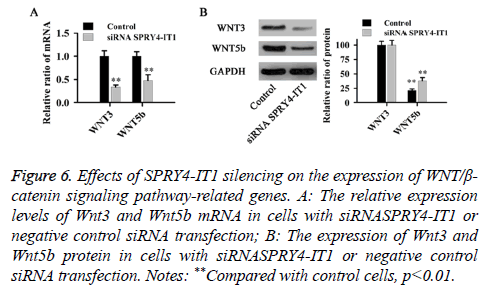
The effect of SPRY4-IT1 on the WNT/β-catenin pathway in OS cells
The relative expression levels of Wnt3 and Wnt5b mRNA and protein were detected by qRT-PCR and western blot, respectively. As shown in Figure 6, compared with control cells, SPRY4-IT1 silencing significantly reduced the expression of Wnt3 and Wnt5b at both mRNA and protein levels (p<0.01).
Figure 6: Effects of SPRY4-IT1 silencing on the expression of WNT/β- catenin signaling pathway-related genes. A: The relative expression levels of Wnt3 and Wnt5b mRNA in cells with siRNASPRY4-IT1 or negative control siRNA transfection; B: The expression of Wnt3 and Wnt5b protein in cells with siRNASPRY4-IT1 or negative control siRNA transfection. Notes: **Compared with control cells, p<0.01.
Discussion
Previous studies have shown that expression level of SPRY4- IT1 was usually upregulated in various cancer tissues [17,18]. Increased expression level of SPRY4-IT1 in cancer cells can increase cell proliferation rate, which in turn promotes the development of cancer [18]. Consistent results were found in our study. After the analysis of SPRY4-ITl expression in OS tissues and normal tissues of 175 patients, we found that expression level of SPRY4-IT1 RNA in OS tissue was significantly higher than that in normal tissue (p<0.05) (Figure 1A). In addition, SPRY4-IT1 was found to be highly expressed in most of the patients with OS (96/157). Our data suggested that expression of SPRY4-IT1 may be positively correlated with the development of OS.
Cyclin D1, which is a nuclear protein, is necessary for cell cycle progression in G1 [19]. It has been reported that expression level of cyclin D1 was usually upregulated in tumor tissues and tumor-derived cell lines [20], and the increased level of cyclin D1 promoted cell division, so as to promote tumor growth [21]. MMP-2 and MMP-9, which are two members of Matrix Metalloproteinase (MMP) family, are responsible for the breakdown of Extracellular Matrix (ECM) in various physiological processes including tumor metastasis [22]. Previous studies have shown that the increased expression levels of MMP-2 and MMP-9 in various cancers were related to the decreased survival rate of the patients [23]. In our study, expression levels of cyclin D1, MMP-2 and MMP-9 were significantly decreased by SPRY4-IT1 silencing at both mRNA and protein levels, indicating that SPRY4-IT1 can positively regulate the expression of cyclin D1, MMP-2 and MMP-9 (Figures 4A and 4B). In addition, MTT assay, colony formation assay and cell migration and invasion assay have shown that growth, colony formation, migration and invasion of OS cells were all reduced by SPRY4-IT1 silencing (Figures 2 and 3). Pearson correlation analysis also showed that the expression of SPRY4-IT1 was positively correlated with the expression of cyclin D1, MMP-2 and MMP-9 (Figure 5). Our data suggested that SPRY4-IT1 can promote tumor growth and metastasis possibly by upregulating cyclin D1, MMP-2 and MMP-9, which are important for cell division and migration.
The WNT/β-catenin signaling pathway has been reported to play pivotal roles in the development of various diseases in both human and animals, and the altered expression of WNT/ β-catenin signaling pathway-related protein can usually lead to the onset of various processes [24]. Wnt3 and Wnt5b are two key players in WNT signaling pathway. Previous studies have shown that WNT3 and WNT5B can promote epithelial cancer cell invasion [25]. Consistent results were found in our study. Cell invasion and migration assay showed that the migration and invasion abilities of OS cells were significantly reduced by SPRY4-IT1 silencing (Figure 3), in addition, SPRY4-IT1 silencing also significantly decreased the expression levels of Wnt3 and Wnt5b at both mRNA and protein levels (Figure 4). Our data suggested that SPRY4-IT1 can promote the migration of OS possibly by upregulating the WNT/β-catenin signaling pathway-related protein Wnt3 and Wnt5b.
In conclusion, SPRY4-IT1 can regulate the growth, migration and invasion of OS cells. These findings suggest that SPRY4- IT1 plays an important role in the regulation of OS progression possibly by upregulating the expression of Cyclin D1, MMP-2, MMP-9 and WNT/β-catenin signaling pathway-related genes. So, SPRY4-IT1 may be a novel biomarker for the prognosis of osteosarcoma.
References
- Lin PP, Patel S. Osteosarcoma. Bone Sarcoma Springer US 2013; 75-97.
- Whelan J, McTiernan A, Cooper N, Wong YK, Francis M. Incidence and survival of malignant bone sarcomas in England 1979-2007. Int J Cancer 2012; 131: E508-517.
- Zhang Y, Zhang L, Zhang G, Li S, Duan J. Osteosarcoma metastasis: prospective role of ezrin. Tumour Biol 2014; 35: 5055-5059.
- Luetke A, Meyers PA, Lewis I. Osteosarcoma treatment where do we stand? A state of the art review. Cancer Treat Rev 2014; 40: 523-532.
- Allison DC, Carney SC, Ahlmann ER, Hendifar A, Chawla S. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma 2012; 2012: 704872.
- Botter SM, Neri D, Fuchs B. Recent advances in osteosarcoma. Curr Opin Pharmacol 2014; 16: 15-23.
- Khandekar MJ, Cohen P, Spiegelman BM. Molecular mechanisms of cancer development in obesity. Nat Rev Cancer 2011; 11: 886-895.
- Chung TT, Pan MS, Kuo CL. Impact of RECK gene polymorphisms and environmental factors on oral cancer susceptibility and clinicopathologic characteristics in Taiwan. Carcinogene 2011; 32: 1063-1068.
- Shafiee R, Javanbakht J, Atyabi N. Diagnosis, classification and grading of canine mammary tumours as a model to study human breast cancer: an Clinico-Cytohistopathological study with environmental factors influencing public health and medicine. Cancer cell Int 2013; 13: 469.
- Vineis P, Wild CP. Global cancer patterns: causes and prevention. Lancet 2014; 383: 549-557.
- Perkel JM. Visiting “non-codarnia”. Biotechniques 2013; 54: 303-304.
- Wang K, Liu F, Zhou LY, Long B, Yuan SM. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 2014; 114: 1377-1388.
- Takahashi K, Yan I, Haga H, Patel T. Long noncoding RNA in liver diseases. Hepatology 2014; 60: 744-753.
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011; 1: 391-407.
- Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol 2014; 35: 7743-7754.
- Peng W, Wu G, Fan H, Wu J, Feng J. Long noncoding RNA SPRY4-IT1 predicts poor patient prognosis and promotes tumorigenesis in gastric cancer. Tumour Biol 2015; 36: 6751-6758.
- Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 2011; 71: 3852-3862.
- Shi Y, Li J, Liu Y, Ding J. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol Cancer 2015; 14: 51.
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev 1993; 7: 812-821.
- Jiang W, Kahn S M, Zhou P. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene 1993; 8: 3447-3457.
- Halilovic E, She QB, Ye Q. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res 2010; 70: 6804-6814.
- Bauvois B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochimica et Biophysica Acta (BBA) Rev Cancer 2012; 1825: 29-36.
- Langers AMJ, Verspaget HW, Hawinkels L. MMP-2 and MMP-9 in normal mucosa are independently associated with outcome of colorectal cancer patients. Br J Cancer 2012; 106: 1495-1498.
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell 2012; 149: 1192-1205.
- Kato S, Hayakawa Y, Sakurai H. Mesenchymal-transitioned cancer cells instigate the invasion of epithelial cancer cells through secretion of WNT3 and WNT5B. Cancer Sci 2014; 105: 281-289.