Research Article - Biomedical Research (2017) Volume 28, Issue 2
Qualitative features selection techniques by profiling statistical features of ECG for classification of heart beats
Chinmay Chandrakar* and Monisha SharmaDepartment of Electronics and Telecommunication, Shri Shankaracharaya College of Engineering and Technology, Bhilai, India
- *Corresponding Author:
- Chinmay Chandrakar
Department of Electronics and Telecommunication
Shri Shankaracharaya College of Engineering and Technology, India
Accepted date: June 01, 2016
Abstract
The measurement of the electrical activity of the heart can be done with electrocardiogram (ECG). Automatic arrhythmia-diagnosis systems which results in high accuracy rates for inside and outside patient are still an important area of research. The accuracy of such system depends on accuracy of the classification system. All this classification system required qualitative features for classification. This paper proposed a unique method of profiling of statistical features for selection of qualitative features through ECG waveform. The proposed approach for selection of qualitative features can classify and differentiate abnormal heartbeats and normal (NORM). Left bundle branch block (LBBB), right bundle branch block (RBBB), ventricular premature contractions (VPC) and atrial premature contractions (APC) comes under abnormal heart beats.
Keywords
ECG waveform, Profiling, Memory elements, Counters.
Introduction
The electrocardiogram (ECG) is graphical representation of the electrical activity of the heart. PQRS complex waveform detection is necessary to determine the heart rate and arrhythmias types such as Tachycardia, Bradycardia and Heart Rate variation. It is also necessary for further processing of the ECG signal in order to detect abnormal beats [1].
After the beat detection, effective beat classification is performed for arrhythmia analysis of normal and abnormal ECG waveform. Maximum likelihood, artificial neural networks and support vector machines have been introduced so far for the ECG beat classification [2,3]. The paper is prepared as follows. In Section II we discuss work related to QRS classification systems. We discuss the working of our signal processing in Section III; qualitative feature selection techniques in Section IV. We discussed our simulation results in Section V. And we conclude our paper in Section VI.
Prior Work
For patient diagnosis QRS complex detection and classification is very important so that one can identify normal and abnormal heart beat. Previous work has been done in the detection of waveform, for example Senhadji et al. who compared the performance of wavelet transform (Daubechies, Spline and Morlet) to identify and explain isolated cardiac beats [4]. Sahambi et al. used a derivative of first order for the classification of the ECG waveforms [5]. The algorithms based on spectral has been developed by author [6,7] or wavelet features [8,9], amplitude activity [10,11] and spatial context [12,13], ECG signal characterization [14-19].
The authors of this paper worked on Z eighty processor for detection of QRS in real time [14]. Ning et al. proposed a technique to determine a exact peak based on largest magnitude within a fixed time window [20]. Jain et al. Proposed, the ECG Feature Extractor provided by LabVIEW Biomedical toolkit detects QRS waves [21]. Yazdani et al. proposed that for QRS detection with peaks at R-waves and valleys before and after, Q and S-points. QRS-onset and QRSoffset are calculated [22]. Nallathambi et al. proposed, the preprocessed ECG signal is converted into a train of pulses using the IF sampler [23].
Beat classification can be required for analysis of arrhythmia type. Methods such as recognition of pattern, likelihood, artificial neural network and vector machines have been extensive used for ECG beat classifications [24-26]. New information can be mapped by continues training using the method of learning the machine [27]. De Chazal et al. classified the beats by analyzing the RR intervals and ECG morphology features along with heart beat segmentation information [28].
Christov et al. proposed a method of classification by measuring the ECG features such as time and frequency [29]. Clinical cardiac signal can be verified using the method proposed in [30] is a patient dependent classifier. Haseena et al. [31] use a combination of fuzzy clustering and artificial neural networks to differentiate between different types of beat class.
Signal Processing of ECG
Pre processing
The presence of unwanted signal causes a big problem in the identification of ECG signals and lots of works have been done in the field to remove these noise signals. By conducting a mathematical method based on varying window length as according to the distance from the adjacent Rpeak which are assumed to be high frequency noise (power line interference, electromyography noise) is removed with the help of the varying window mean procedure form by the universal time constant equation and the low frequency noise (base-line wandering, motion artifact) removal task is done with the help of Fast Fourier Transform [32].
QRS detection and extraction of features
The QRS complex is the important waveform in the electrocardiogram (ECG). QRS detection provides the basic for all ECG analysis algorithms which has been done automatically. We detected the QRS complex and QRS features (‘‘H-QR”, ‘‘H-RS”, ‘‘QRS-duration”, ‘‘Slope-R”, ‘‘Slope-RS” and ‘‘Ratio-RR”) by using Lab View as proposed in paper [33].
Qualitative Features Selection Techniques
Profiling approach using counters
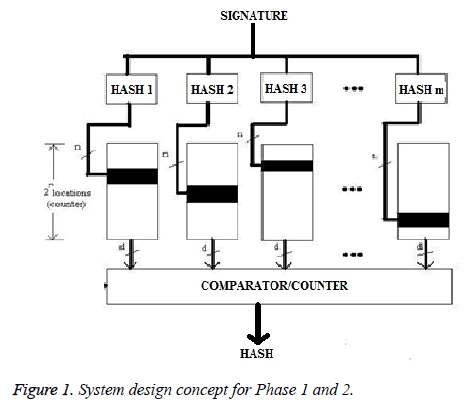
The basic concept in ECG profiling is based on the repetitionbased pattern detection schemes, to derive a normal and abnormal (LBBB, RBBB, VPC and APC) ECG pattern for an individual. Our proposed method for profiling is based on repetitions of pattern as shown in Figure 1. The structure of these repeated parameters is specific (different class of arrhythmia) and depend to the features of each behavior. Therefore the proposed system is well synchronized to particular class so that the output will be a profiling curve for specific application. In our system, the data information chosen is morphology of for QRS complex and the ratio of the present and the past R peak points.
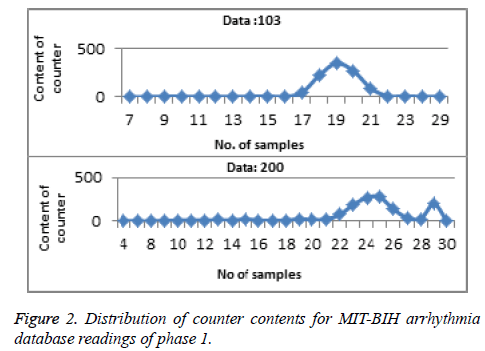
Phase 1: In Phase 1, once the QRS complex and features, different statistical information that is in each QRS waveform (‘‘H-QR”,‘‘H-RS”, ‘‘QRS-dur”, ‘‘Slope-R”, ‘‘Slope-RS” and ‘‘Ratio-RR”) has to be determined for the particular class of arrhythmia than we plotted the profile curve by using the feature (QRS-duration, information in form of number of samples). Depending on the information of the counters, a hyperbolic profiling curve would be obtained, where abnormality should be found on the end of the curve [24]. Abnormal ECG beats of the ECG profiling curve will be seen where humps exist on the tail of the bell-shaped curve. If no anomalies lie on the tail(s) than we declare that ECG waveform belong to normal class and on the other hand if anomalies found on the end of the curve than we declare that the waveform is having an abnormal ECG beats as shown in Figure 2 and then now onwards remaining features act as a signature for phase 1.
Phase 2: In Phase 2, m independent hash values for phase 2 are used from the phase 1 signature. These hash values is used as memory indices in phase 2. Hash functions were designed similar to phase 1. The array i has been indexed by i. The location of the repeated value is incremented hence the 2n locations of the memory also function as counters. The counters chosen by hash units will be incremented. After calculating final result for phase 1, the ‘m’ independent hash feature for phase 2 has been determined. The individual features will act as an independent hash feature i.e. m. This process is repeated for all the class of heart beat. Due to the time-varying morphology of the signal subject to physiological conditions there is a variation in QRS waveform (duration and amplitude), hence the values of the features of different class. Once the hash feature has been found for phase 2, then for the respective hash feature of the entire class of waveform for less than one minute, the values were determined. This value was used to index into array i. The resultant value acts as a memory address ‘n’ for the array i. Therefore when hit location is incremented, the array locations also function as a counter.
Counter index in the respective memory elements is actually the hash result of the ECG features (‘‘H-QR”, ‘‘H-RS”, ‘‘Slope-R”, ‘‘Slope-RS” and ‘‘Ratio-RR”) which also indicate to the location of memory. This is a data for classifying the ECG features. Each location of memory is also a counter; i.e., whenever a hashed value of ECG feature is generated, the data of that position is incremented by one. The profile curve for the respective feature (results from the respective memory element) has to be plotted. The counter content x distribution will be a normal (bell-shaped curve) or abnormal (not bellshaped curve). The features which result in the bell-shaped will be selected as a qualitative feature for discriminating the heartbeat of that particular class. We paired those features which result in the bell-shape. Now onward these paired features will act as a signature of phase 2, for classifying the waveform of particular class. Similarly the waveform of different class of heart beats or group of different class of heart beat from the MIT-BIH Arrhythmia are taken for determining the qualitative features of respective class as discussed above, so that the system can be well trained for that particular heartbeat class. We tuned our system for classifying the waveform of ten arrhythmia classes of NORM, LBBB, RBBB, VPC, APC, NORM-APC, LBBB-VPC and RBBB-APC etc. the waveform of ten arrhythmia classes of NORM, LBBB, RBBB, VPC, APC, NORM-APC, LBBB-VPC and RBBBAPC etc.
Profiling approach using joint and disjoint
The process of qualitative features selection (QFS) can be done after determining the statistical features of ECG. Let Ri,j indicates value range of the ith complex feature for the jth beat case. The sub-index i is defined as the same as that of Fi. The sub-index j=1, 2, 3, 4, 5 denotes the case of ‘‘normal”, ‘‘lbbb”, ‘‘rbbb”, ‘‘vpc” and ‘‘apc”, respectively.
The following steps are followed to obtain QFS:
Step 1: Defining the variables for feature and heartbeat case:x
Let k=Heartbeat case variable for comparing various heartbeats where k ≠ j. Ri,j=It is the value of feature which ranges from ith ECG complex feature for jth beat case. Ri,k=It is the feature valve range of jth PQRST complex feature for kth heartbeat case (Tables 1 and 2).
| Value of i | Feature represented by i |
|---|---|
| 1 | H-QR |
| 2 | H-RS |
| 3 | QRS-duration |
| 4 | QTP-interval |
| 5 | Ratio-RR |
| 6 | Slope-QR |
| 7 | Slope-RS |
| 8 | Area-QRS |
| 9 | Area-R’ST’ |
Table 1. PQRST complex feature.
| Value of j and k | Cases represented by j and k |
|---|---|
| 1 | NORMAL |
| 2 | LBBB |
| 3 | RBBB |
| 4 | VPC |
| 5 | APC |
Table 2. Heartbeat case.
i=PQRST complex feature and 1≤ i ≤ 9.
j, k=heartbeat case j ≠ k 1 ≤ j ≤ 5 , 1 ≤ k ≤ 5
Step 2: Comparison between 2 heartbeat cases:
Two heartbeat cases are compared to find out if any particular feature can discriminate two heartbeat cases or not.
If R=Rij ∩ Rik, Then,
Case-1: Rij ∩ Rik =1, Ri,k and Ri,j are the features value and if they do not overlap. Two heartbeat cases (j and k) will not coincide. Hence, it can be concluded that the feature ‘i’ can discriminate heartbeat cases j and k.
Case-2: Rij ∩ Rik =0, The heartbeats case-k and case-j cannot be discriminated and Fi is not obtained if Ri,k and Ri,j overlap . Two heartbeat cases (j and k) will coincide.
Step 3: Features that can discriminate two heartbeat cases have been founded:
Let Fi = feature that can discriminate j and k heartbeat cases and NFi=Total no. of features that can discriminate j and k heartbeat cases.
Step 4: Arrange NFi, i=1, 2, . . . , 9, in the descending order values and then highest value index will be selected, that is, i=argument (maximum{NFi}, i=1,2,3,…,9)
Step 5: Fi qualitative feature can be obtained
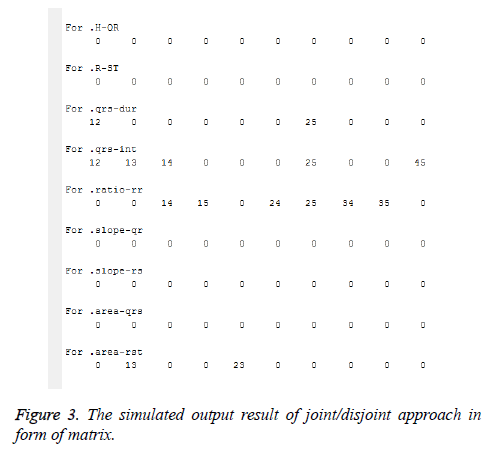
Step 6: End. QRS-duration, QTP-interval, Ratio-RR and Area- RST as qualitative features were selected after running the above algorithm as shown in Figure 3. Each heartbeat case had its own range of values for each qualitative feature.
Let us assume a, b, c and d as QRS-dur., QTP-int, Ratio-RR and Area-R’ST’ respectively. The complete set of heartbeat cases from the matrix is represented by 1, 2, 3...9, 10 and qualitative features are represented by a, b, c and d and it can be written in the form of set as: aUbUcUd ={1,2,3,4,5,6,7,8,9,10}. This represents the complete set which is formed by the intersection of all the four qualitative features which contains ten different heartbeat cases combination.
a ε [1,2]
This represents that the qualitative feature ‘a’ forms a set which comprises of heartbeat cases combination 1, 2.
b ε [1,2,3,4,5]
This represents that the qualitative feature ‘b’ forms a set which comprises of heartbeat cases combination 1, 2,3,4,5.
c ε [2,4,6,7,8,9]
This represents that the qualitative feature ‘c’ forms a set which comprises of heartbeat cases combination 2, 4 and 6,7,8,9.
d ε [3,10]
This represents that the qualitative feature ‘d’ forms a set which comprises of heartbeat cases combination 3, 10.
Simulation Results
We have applied our qualitative features selection techniques on the record of arrhythmia database [3]. NORM, LBBB, RBBB, VPC and APC are abbreviation used for each heart ‘‘beat”, which contain different arrhythmia in the signal at different time instance. This simulation result has taken three data of the NORM case: Tape #222 (2274 NORM beats), #223 (2230 NORM beats) and #234 (2700 NORM beats). The abnormal heart ECG signals, records are: (1) two records of the left bundle branch block (LBBB) case (Tape #111 (2123 LBBB beats) and #214 (2002 LBBB beats)); (2) two records of the right bundle branch block (RBBB) case (Tape #118 (2166 RBBB beats) and #231 (562 RBBB beats)); (3) three records of ventricular premature contractions (VPC) case (Tape #200 (826 VPC beats), #221 (396 VPC beats) and #233 (831 VPC beats)); and (4) two records of atrial premature contractions (APC) case (Tape #222 (209 APC beats) and #232 (1382 APC beats)).
Simulation result of cardiac profiling approach
The features which result in the bell-shaped will be selected as a qualitative feature for discriminating the heartbeat of that particular class as discuss in section IV (A). We paired those features which result in the bell-shape so that the system can be fine-tuned for that particular class. Thus we obtain the heart beat cases being discriminated by the different features (Table 3).
| HEART BEAT CASES | QUALITATIVE FEATURES |
|---|---|
| NORMAL | QRS-DUR |
| LBBB | H-QR,H-RS,SLOPE-QR,SLOPE-RS |
| RBBB | H-RS,SLOPE-QR,SLOPE-RS,RATIO -RR |
| VPC | H-QR, SLOPE-QR,SLOPE-RS, RATIO -RR |
| APC | H-QR |
| NORMAL -RBBB | H-QR,H-RS |
| NORMAL-VPC | H-QR, SLOPE-QR,SLOPE-RS, RATIO -RR |
| NORMAL-APC | H-QR, RATIO -RR |
| LBBB-VPC | H-QR,H-RS,SLOPE-QR,SLOPE-RS, RATIO -RR |
| RBBB-APC | H-RS,SLOPE-QR,SLOPE-RS,RATIO -RR |
Table 3. Heart beat cases discriminated by the different features resulting in the bell-shape.
Simulation result of joint and disjoint approach
From Figure 3 pair of cases which was found in the respective qualitative features and can be written as:
aUbUcUd={12,13,14,15,23,24,25,34,35,45}, a ∩ b={12}, a ∩ c={14}, a ∩ d={13}, a ∩ b ∩ c={25}, 12 ? {a,b}, 13 ? {a,d}, 14 ? {a,c}, 15 ? {c}, 23 ? {d}, 24 ?{c}, 25 ? {a,b,c}, 34 ? {c}, 35 ? {c} and 45 ? {a}.
Thus we obtain the heart beat cases being discriminated by the different features (Table 4).
| HEART BEAT CASES | QUALITATIVE FEATURES |
|---|---|
| NORMAL- LBBB (12) | QTP-INT,QRS-DUR |
| NORMAL -RBBB (13) | QTP-INT,AREA-RST |
| NORMAL-VPC (14) | QTP-INT,RATIO-RR |
| NORMAL-APC(15) | RATIO-RR |
| LBBB-RBBB(23) | AREA-RST |
| LBBB-VPC(24) | RATIO-RR |
| LBBB-APC(25) | QRS-DUR,QTP-INT,RATIO-RR |
| RBBB-VPC(34) | RATIO-RR |
| RBBB-APC(35) | RATIO-RR |
| VPC-APC(45) | QTP-INT |
Table 4. Heart beat cases being discriminated by the different features.
Conclusion
We proposed a novel technique for extraction of qualitative features used for classification of ECG waveform. Unlike other training classification methods, we extracted qualitative features with different sample of arrhythmia types and also with samples of normal beats, so that our system can differentiate between arrhythmia of different types. The raw data used for qualitative features extraction system is large number of almost abnormal beats. Otherwise, if we trained our system with only normal beats, the system cannot verify the beats as abnormal; it is due to abnormal behaviour now become the normal. As ECG is a non-stationary signal i.e. morphology of ECG signal depends on physical conditions of person, it varies with time and it differs for person to person, therefore out of both the proposed approach profiling approach using counters is much more suitable for classification because it depends on packets of data stream for extraction of qualitative features as compared to joint/disjoint approach which depends on maximum and minimum value of individual features. Extracted qualitative features can able to differentiate different types of arrhythmia and can be used in classification system for classification of different types of arrhythmia.
References
- Lilly LS. Pathophysiology of heart disease, 3rd edition. Lippincott Williams and Wilkins, Philadelphia, 2003, 57-90.
- SNORT Network Intrusion Detection System, www.snort.org.
- MIT-BIH (www.physionet.org)
- Senhadji L, Carrault G. Comparing wavelet transforms for recognizing cardiac patterns. IEEE Eng Med Biol 1995; 14: 167-173.
- Sahambi JS, Tandon SM, Bhatt RKP. Using wavelet transforms for ECG characterization: an on-line digital signal processing system. IEEE Eng Med Biol 1997; 16: 77-83.
- Hamilton PS, Tompkins WJ. Quantitative investigation of QRS detection rules using the MIT-BIH arrhythmia database. IEEE Trans Biomed Eng 1986; 33: 1157-1165.
- Thakor NV, Webster JG, Tompkins WJ. Estimation of QRS complex power spectra for design of a QRS filter. IEEE Trans Biomed Eng 1986; 31: 702-706.
- Li C, Zheng C, Tai C. Detection of ECG characteristic points using wavelet transforms. IEEE Trans Biomed Eng 1995; 42: 21-28.
- Castro B, Kogan D, Geva AB. ECG feature extraction using optimal mother wavelet. IEEE EMBE Int Conf 2000.
- Romero I, Serrano L. ECG frequency domain features extraction: A new characteristic for arrhythmias classification. IEEE Eng Med Boil Soc 2001.
- Kadambe S, Murray R, Boudreaux-Bartels GF. Wavelet transform based QRS complex detector. IEEE Trans Biomed Eng 1999; 46: 838-848.
- Romero L, Addison PS, Grubb N. R-wave detection using continuous wavelet modulus maxima. IEEE Proc Comp Cardiol 2003; 30: 565-568.
- Tkacz EJ, Komorowski D. An improved statistical approach to the QRS detection problem using matched filter facilities. Biomed Eng 1992; 37: 99-109.
- Pan J, Tompkins WJ. A real-time QRS detection algorithm. IEEE Trans Biomed Eng 1985; 32: 230-236.
- Dotsinsky IA, Stoyanov TV. Ventricular beat detection in single channel electrocardiograms. Biomed Eng Online 2004; 3: 3.
- Zhang F, Tan J, Lian Y. An effective QRS detection algorithm for wearable ECG in body area network. IEEE Biomed Circuits Syst Conf 2007.
- Braojos R, Beretta I, Ansaloni G, Atienza D. Early classification of pathological heartbeats on wireless body sensor nodes. Sensors 2014; 14: 22532-22551.
- Zhou HY, Hou KM. Embedded real-time QRS detection algorithm for pervasive cardiac care system. 9th IEEE Int Conf Signal Process 2008.
- Cvikl M, Zemva A. FPGA-oriented HW/SW implementation of ECG beat detection and classification algorithm. Digital Signal Process 2010; 20: 238-248.
- Ning X, Selesnick IW. ECG Enhancement and QRS Detection Based on Sparse Derivatives. Biomed Signal Process Control 2013; 8: 713-723.
- Jain S, Kumar P, Subashini MM. LABVIEW based expert system for Detection of heart abnormalities. International Conference on Advances in Electrical Engineering (ICAEE), 2014.
- Yazdani S, Vesin J-M. Adaptive Mathematical Morphology for QRS Fiducial Points Detection in the ECG. Comput Cardiol 2014; 38: 725-728.
- Nallathambi G, Principe JC. Integrate and Fire Pulse Train Automaton for QRS detection. IEEE Transactions Biomed Eng 2014; 61: 317-326.
- Faezipour M, Nourani M, Panigrahy R. A real-time worm outbreakdetection system using shared counters. 15th Annual IEEE Symp High Perform Interconnects 2007.
- Song MH, Lee J, Cho SP, Lee KJ, Yoo SK. Support vector machine based arrhythmia classification using reduced features. Int JControl Autom Syst 2005; 3: 571-579.
- Besrour R, Lachiri Z, Ellouze N. ECG beat classifier using support vector machine. 3rd IEEE Int Conf Inf Commun Technol 2008.
- Hu YH, Palreddy S, Tompkins WJ. A patient adaptive ECG beat classifier using a mixture of experts approach. IEEE Trans Biomed Eng 44: 891-900.
- de Chazal P, O’Dwyer M, Reilly RB. Automatic classification of heartbeats using ECG morphology and heartbeat interval features. IEEE Trans Biomed Eng 2004; 51: 1196-1206.
- Christov I, G´omez-Herrero G, Krasteva V, Jekova I, Gotchev A, Egiazarian K. Comparative study of morphological and time-frequency ECG descriptors for heartbeat classification. Med Eng Phys 2006; 28: 876-887.
- Iliev I, Krasteva V, Tabakov S. Real-time detection of pathological cardiac events in the electrocardiogram. Physiol Meas 2007; 28: 259-276.
- Haseena HH, Mathew AT, Paul JK. Fuzzy clustered probabilistic and multi layered feed forward neural networks for electrocardiogram arrhythmia classification. J Med Syst 2009.
- Chandrakar C, Sharma M. A real time approach for ECG signal denoising and smoothing using adaptive window technique. IEEE 9th International Conference on Industrial and Information Systems (ICIIS), 2014.
- Chandrakar C, Sharma M. A novel mathematical ECG signal analysis approach for features extraction using labview. J Software Eng 2014; 8: 1-7.