Research Article - Current Pediatric Research (2017) Volume 21, Issue 1
Mutational analysis of NPHS2 and WT1 genes in Saudi children with nephrotic syndrome.
Abdulla A Alharthi1,2,3, Ahmed Gaber1,4, Mohamed W AbuKhatwah3, Abeer M Almalki5, Abdullah A Muzallef6, Mohamed M Hassan1,7, Ehab I El-Hallous1,8, Meshari M Dalbouh6, Gadah H Ali6, Hanan M Atyah61Deanship of Scientific Research, Taif University, Taif, KSA.
2Faculty of Medicine, Department of Pediatrics, Taif University, Taif, KSA.
3Pediatrics Department, Alhada Armed Forces Hospital, Taif, KSA.
4Department of Genetics, Faculty of Agriculture, Cairo University, Giza, Egypt.
5Children Hospital, Taif, KSA.
6Abha Maternity Children Hospital, Abha, KSA.
7Genetics Department, Faculty of Agriculture, Minufiya University, Egypt.
8Department of Zoology, Faculty of Science, Al-Arish, Suez Canal University, Egypt.
- *Corresponding Author:
- Abdulla A Alharthi
Deanship of Scientific Research
Faculty of Medicine, Taif University
P.O. Box 689, Zip code 21944, Taif
Kingdom of Saudi Arabia.
Tel: 0966505760013
Fax: 00966127274299
E-mail: aharthy@gmail.com
Accepted date: November 25, 2016
Abstract
Background: Nephrotic syndrome is the predominant glomeurular disease in childhood. Mutations in numerous genes are known to be the reason for steroid-resistant nephrotic syndrome; however, the presence of these mutations seems to be effected by race sociocultural differences and interethnic group. Mutations in NPHS2 and WT1 genes record for nearly 20% and 5% of all children cases with steroid-resistant nephrotic syndrome, respectively. By contrast, mutations are absent from children with either steroid-dependent nephrotic syndrome or frequently-relapse nephrotic syndrome.
Methods: Mutation analysis was accomplished by direct sequencing of the complete 8 exons of NPHS2 and exons 8 and 9 of WT1 in 20 patients with steroid-resistant nephrotic syndrome, 25 with steroid-dependent nephrotic syndrome, and 13 with frequently-relapse nephrotic syndrome.
Results: Three pathogenic mutations in NPHS2 were detected within steroid-resistant nephrotic syndrome patients. One as a non-sense mutation in exon 1 that was reported previously, while the second was novel and found as missense mutation in exon 7. The third one was found in the NPHS2 promoter region. Additionally, for the first time, one pathogenic missense mutation in exon 8 of WT1 gene was found in one Saudi patient with steroid-resistant nephrotic syndrome. All four mutations were documented and submitted to the ClinVar database.
Conclusion: NPHS2 and WT1 genes mutations are risk factors for steroid-resistant nephrotic syndrome with about 15% and 5% in Saudi pediatric patients. More molecular studies are required to clarify other possibility genes that responsible for the development of steroidresistant nephrotic syndrome in Saudi children.
Keywords
Steroid-resistant nephrotic syndrome, Steroid-dependent nephrotic syndrome, Pediatric, WT1, NPHS2, Saudi Arabia.
Introduction
Nephrotic Syndrome (NS) is defined as the occurrence of proteinuria, hypoalbuminemia, edema, and hyperlipidemia and it represents one of the most serious diagnoses made in pediatric nephrology [1]. The appearance of NS may be as a result of primary (idiopathic) glomerular or secondary (systemic) disease [2]. According to the response of the patients to the steroid therapy, NS has been divided into two different categories: Steroid-Sensitive Nephrotic Syndrome (SSNS) versus Steroid-Resistant Nephrotic Syndrome (SRNS) [3]. A bout 70% of SSNS patients will have one or more relapses, and a significant percentage will go on to develop either Frequent-Relapsing NS (FRNS) or steroid-dependent NS course (SDNS) [2].
It is a well-known that SRNS is the familiar phenotype of the genetically forms of NS whether recessive or dominant [4]. More recent report indicates that the occurrence of single-gene causation of SRNS can be found in at least a one-third of all tested families [5]. Up to date, several causative genes related to NS have been identified by either using direct DNA sequencing approaches or next-generation sequencing technology [6-12]. From literature, NPHS1, NPHS2 and WT1 are the most analyzed genes especially in a large cohort of patients with NS [3,9,13]. The results proposed that, NPHS1 and WT1 mutations were essentially found in children with congenital NS, while NPHS2 mutations might be more prevalent in children with idiopathic SRNS [3,13]. The age of onset for NPHS2 mutation is varying but commonly before 6 years of age [14]. By contrast, adult-onset before 18 years old cases of SRNS related with NPHS2 are also well documented, these mutations are often found to be together as a compound heterozygous state [15].
Mutations in NPHS2 and WT1 genes have been reported to contribute nearly to 20% and 5% of all cases of SRNS, respectively [3,9,16]. On the other hand, NPHS2 and WT1 gene mutations are absent from children with SSNS, even though, all SSNS patients are not clinically same group as some may be FRNS or SDNS [17]. Until now, the influence of NPHS2 and WT1 mutations in FRNS or SDNS patients is fully unknown [17]. It was suggested that children with heterozygous mutations in NPHS2 gene may have middle disease course between SSNS and SRNS that displays as the FRNS/SDNS course, but this finding did not report in children with uncomplicated SSNS [18].
With the growing number of undiagnosed missense mutations, many diagnostic laboratories use in silico prediction tools, e.g. sequence and evolutionary conservation-based tools and protein sequence and structure-based tools, to anticipate the effect the pathogenicity of a novel mutation in relation to the evolutionary conservation of specific amino acids, as well as protein structure and function [19]. Some of these tools are used frequently by diagnostic labs to recommend clinicians of disease probability in the absence of previous indication [19,20].
The population of the Kingdom of Saudi Arabia (KSA) is categorized with high consanguinity rate (52%-56% of marriages), therefore, it is documented that the high frequency of autosomal recessive genetically mediated renal diseases is due to the high consanguinity rate [21,22]. Also, it is well recognized that NS is more common in Asian children than in Caucasian children [23]. Furthermore, congenital and infantile NS was also reported to be higher in KSA than in other countries [22,24].
Therefore, in the present study, we performed mutational analysis of the entire exons of NPHS2 plus exons 8 and 9 of WT1 genes in 20 idiopathic cases of SRNS, 25 with SDNS and 13 cases with FRNS.
Materials and Methods
Patients
We identified all children with a clinical diagnosis of NS aged between 1-16 years old who were being followed at different Children Hospitals in western area of KSA under parent's consent and approved protocols of the central Hospital Bioethics committee of Alhada armed forces hospital under number (H-02-T-001-PTRC-15-04-226). NS patients were entered to the hospital from January, 2012 to December 2015. NS patients was defined as proteinuria (>40 mg/m2/h), hypoalbuminemia, and generalized edema [25]. SRNS patients were defined as the failure to response to daily therapy of prednisone (2 mg/kg) for 4-6 weeks [17,25]. Congenital Nephrotic Syndrome (CNS) was defined as the presentation of NS within the first three months of life [25]. FRNS patients defined as two or more relapses during the first 6 months or more than three relapses during any 12 month period, while, SDNS defined as two successive relapses at the time of tapering period of prednisone therapy or relapse within 2 weeks of the discontinuation of steroid therapy [25]. Inclusion criteria was made up of all Saudi patients with nephrotic syndrome of children below 16 years, while, exclusion criteria was patients with NS of a known secondary cause (e.g. IgA nephropathy). Familial and sporadic NS types were also determined by family history. Renal pathologists assessed renal biopsy specimens. Characteristic features of the clinical diagnosis were documented such as: name, sex, family history, HTN, hematuria, age of onset, response to steroid therapy, histological features of kidney biopsy, and interval time of progression toward End Stage Renal Disease (ESRD).
Genomic DNA Extraction
Genomic DNA was extracted directly from blood according to instructions of genomic DNA extraction kit (Thermo Fisher; USA). Quality of extracted DNA was assessed via electrophoresis on agarose gel and concentration of DNA was estimated by UV Spectrophotometry.
Amplification of DNA via PCR
All eight exons of NPHS2 and exons 8 and 9 of WT1 were amplified from genomic DNA by polymerase chain reaction (PCR) and directly sequenced. All PCR primers were designed from intronic sequences. The sequences of the forward and reverse primers, PCR conditions and the sizes of PCR products are given in Table 1.
Detection of NPHS2 Gene Mutations and Variants by Direct DNA Sequencing
Sequencing of the purified specific fragments using the same primers were employed in the PCR amplification process on both forward and reverse directions and it was repeated to confirm reproducible results. The products were sequenced using the Big Dye Terminator Cycle Sequencing Ready Reaction Kit (ABI Applied Biosystems, Life Technologies) on a 3130 Genetic Analyzer (Applied Biosystems) and the raw sequencing results were collected using the Data collection software version 3.1 from ABI Applied Biosystems. The DNA sequences were analyzed using the Seqscape software version 2.7 from ABI Applied Biosystems for base-calling and mutation detection. The results was compared against the references DNA sequence of NPHS2 (GenBank; NG_007535) and WT1 (GenBank; NG_009272).
In Silico Analysis of the Nucleotide Changes Identified
To assess the possible effect of the pathogenic mutations that found in the present study, three software programs were used: (1) PolyPhen2 (Polymorphism Phenotyping 2) (http://genetics.bwh.harvard.edu/pph2/), which expects possible effect of the non-synonymous exonic nucleotide change in the function of the human proteins [26]. (2) Sorting Tolerant from Intolerant (SIFT) and PROVEAN program (http://provean.jcvi.org/seq_submit.php). PROVEAN is a new calculation method which works for both single nucleotide polymorphisms (SNPs) and/or indels [27]. In PROVEAN program -2.5 is consider as a default threshold, therefore, variants with a score equal to or below -2.5 are considered deleterious, while, variants with a score above -2.5 are considered neutral [27]. (3) The RWebLogo program (plotting custom sequence logos. R package version 1.0.3.) (https://CRAN.R-project.org/ package=RWebLogo), which is a statistical method for detecting amino acid conservation [28]. From information theory, conservation can be calculated at each amino acid position that ranges from zero to 4.3 bits at an equally position for by all the 20 amino acids at an invariant position [29]. Therefore, strongly conserved positions are expected to be more deleterious when substitute with other amino acid, whereas weakly conserved positions are anticipated to tolerate more substitutions. Additionally, we used ConSite program (http://consite.genereg.net) for searching of any transcription factor binding site at the position of the variant that was identified in the promoter region.
SNP Database Submission
The mutations and novel polymorphisms variants that detected in this study were documented and submitted to the ClinVar under the accession numbers SCV000265969, SCV000265972, SCV000265973 and SCV000266491.
Results
We selected 58 Saudi pediatric patients with a diagnosis of NS who visited different hospitals from the western area of KSA during the past 3 years. These selections were including 25 patients (43%) with SDNS, 13 patients (22%) with FRNS, and 20 patients (35%) with SRNS. The mean age at the onset of the NS is 4.5 years while the mean age for the total years is 7.8 years. The majority was boys (55%). 33% of the patients had a positive family history of NS.
| Gender | Gene Mutation | Age of onseta | Renal Biopsyb |
Age at ESRDc | HPTd | Hematuria | Site of mutation | Nucleotide change | Effect on coding |
|
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | NPHS2 | 9 y | FSGS | 12 y | Yes | Microscopic | Promoter region | [c.-52C>G(;) c.-51G>T] | - |
| 2 | Female | NPHS2 | 5 y | FSGS | 7 y | Yes | Microscopic | Exon 1 | c.115C>T | p.Q39X |
| 3 | Female | NPHS2 | 6 y | FSGS | 9 y | Yes | Macroscopic | Exon 7 | c.812C>T | p.P271L |
| 4 | Female | WT1 | 3 m | DMS | 1 y | Yes | Negative | Exon 8 | c.1301G>A | p.R434H |
a) y: years; m: months; b) FSGS: Focal Segmental Glomerulosclerosis; DMS: Diffuse Mesangial Sclerosis; c) ESRD: End-Stage Renal Disease; d) HPT: Hypertension
Table 2. Clinical data of the individual cases with mutations identified in SRNS patients associated with NPHS2 and WT1 genes.
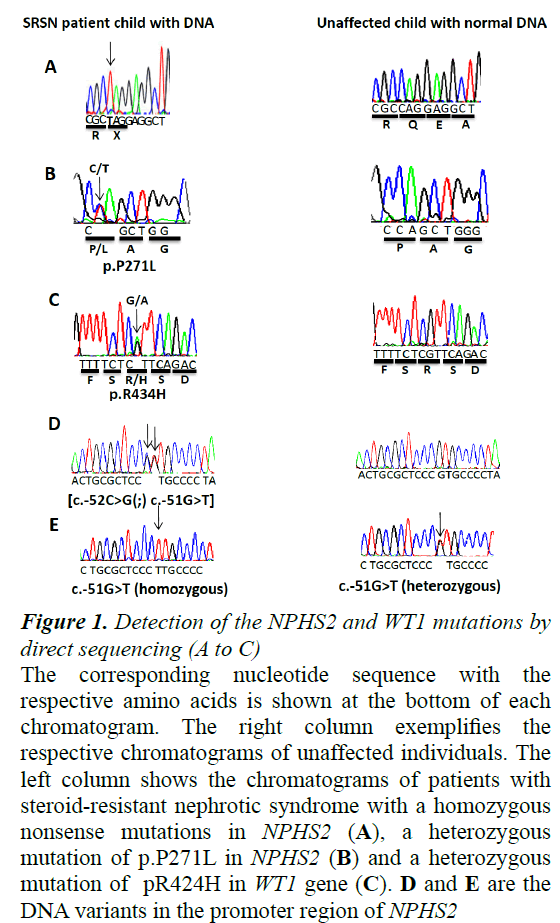
The nomenclature for describing the sequence variations that used here was based on the reference sequence of GENBANK database as NM_014625, AJ279254, and NP_055440 for NPHS2 gene, while NM_024426.3, NP_077744.3 for WT1 gene. A summary of results from the mutation analysis of NPHS2 and WT1 genes is given in Table 2. DNA sequence analysis of all 8 exons of NPHS2 gene revealed 3 pathogenic mutations (3/20; 15%) that were found in SRNS cases only (Figure 1). First mutation was found as homozygous mutation in one girl at 5 years old and located in exon 1 (c.115C>T; p.Q39X). Two years later, this girl had developed to ESRD. This nonsense mutation was reported previously [9]. The second pathogenic mutation (c.812C>T; p.P271L) was found in female patient aged 6 years and diagnosed as SRNS with focal segmental glomerulosclerosis (FSGS) pattern and had macroscopic hematuria (Table 2). This missense mutation is novel and found in exon 7 as a heterozygous state (Figure 1). These two mutations were documented and submitted to the ClinVar database under the accession numbers SCV000265972 and SCV000265973, respectively.
Figure 1. Detection of the NPHS2 and WT1 mutations by direct sequencing (A to C)
The corresponding nucleotide sequence with the respective amino acids is shown at the bottom of each chromatogram. The right column exemplifies the respective chromatograms of unaffected individuals. The left column shows the chromatograms of patients with steroid-resistant nephrotic syndrome with a homozygous nonsense mutations in NPHS2 (A), a heterozygous mutation of p.P271L in NPHS2 (B) and a heterozygous mutation of pR424H in WT1 gene (C). D and E are the DNA variants in the promoter region of NPHS2.
Interestingly, we found one SRNA girl at age 9 years with hypertension and microscopic hematuria having a haplotype or compound heterozygous mutation located side by side [c.-52C>G(;) c.-51G>T] in the binding site of Upstream Stimulatory Factor (USF) of the NPHS2 promoter region (Figure 1). Her renal biopsy was demonstrated as FSGS pattern. Three years later, she had developed ESRD (Table 2). Unfortunately, we could not screen the DNA of the parents for these specific mutations to explore if these variant are found on one chromosome and thus represent as a haplotype heterozygous mutation or they are present on two separate chromosomes and thus the genotype will be a compound heterozygous mutation. This mutation was submitted to the CLinVar under the accession number SCV000266491.
By contrast to these results, no pathogenic homozygous or heterozygous mutations were found in any SDNS or FRNS cases. But, we detected known and novel Single Nucleotide Polymorphisms (SNPs) in some patients with SDNS, FRNS and SRNS. Total seven SNPs variation in NPHS2 gene was detected (Table 3). In the promoter region, one reported SNP was found (c.-51G>T) as heterozygous state in 11 patients with SDNS or FRNS; while, as a homozygous state in 3 patients with SDNS (Table 3 and Figure 1). In exon 1, one silent homozygous mutation (c.102A>G; p.Arg34Arg) was found in 25 patients (Table 3). Exon 8 had five SNPs; c.954 C>T (as homozygous state in 12 patients and heterozygous state in 17 patients), c.*157G>A (as homozygous state in 30 patients), c.*200G>A (as homozygous state in 15 patients and heterozygous state in 6 patients), c.*258A>G (as homozygous state in 2 patients and as heterozygous state in 2 patients), and finally c.*428A>G was found as a homozygous state in 20 patients (Table 3).
| Gene | Exon | Polymorphism | Heterozygous/Homozygous | Patients (n=58) | Effect on coding |
|---|---|---|---|---|---|
| NPHS2 | Promoter region | c.-51G>T | Homozygous Heterozygous |
3 (5%) 11 (20%) |
- |
| 1 | c.102A>G | Homozygous | 25 (43%) | p.Arg34Arg | |
| 8 | c.954C>T | Homozygous Heterozygous |
12 (20.6%) 17 (29%) |
p.Ala318Ala | |
| 8 | c.*157G>A | Homozygous | 30 (52%) | - | |
| 8 | c.*200G>A | Homozygous Heterozygous |
15 (26%) 6 (10%) |
- | |
| 8 | c.*258A>G | Homozygous Heterozygous |
2 (3.5%) 2 (3.5%) |
- | |
| 8 | c.*428A>G | Homozygous | 20 (34.5%) | - | |
| WT1 | Intronic | c.1250-52G>T | Heterozygous | 2 (3.5%) | - |
| Intronic | c.1250-32C>A | Heterozygous | 2 (3.5%) | - |
Table 3. NPHS2 and WT1 polymorphisms in 58 patients with SDNS, FRNS, and SRNS.
When we screened exons 8 and 9 of WT1 gene for possible mutation, we found one female SRNS patient (1/20; 5%) identified as a congenital nephrotic syndrome carrying a heterozygous mutation in exon 8 at the position c.1301G>A (p.R434H) (Figure 1). This mutation was documented and submitted to the ClinVar database under the record SCV000265969. Her biopsy was confirmed with diffuse mesangial sclerosis (DMS). At one year old this patient progressed to ESRD.
Also, we did not detect any disease-causing mutation correlated with WT1 gene in patients with SDNS or FRNS. While, two intronic SNPs (c.1250-32C>A) and (c.1250- 52G>T) were found in WT1 gene in heterozygous state with 2 patients (3.5%) for each SNP (Table 3).
Based on in-silico evaluation for the two heterozygous mutations that found in NPHS2 and WT1 gene by using the web-based programs SIFT/PROVEAN, both mutations of P271L in NPHS2 and R434H in WT1 genes found to be affected the function of the two proteins with a deleterious score of -9.46 and -4.57 for NPHS2 and WT1 proteins, respectively. This prediction is further supported by PolyPhen2 program, which also predicted that the amino acid substitution is probably damaging to the proteins. These results were in agreement with the distribution of the conserved amino acids along the sequences of NPHS2 and WT1 proteins. The RWebLogo program was used to measure the correlation between sequence conservation and tolerance to substitutions [28]. As shown in Figure 2, the two amino acids (P in NPHS2 and R in WT1 proteins) are shown to be well conserved among NPHS2 and WT1 proteins with more than 2 bits, respectively. Therefore the mutations of p.P271L in NPHS2 and p.R434H in WT1 might be a deleterious within the SRNS patients.
Discussion
Although NPHS2 gene mutations cause autosomal recessive familial SRNS, it is also possible that heterozygous mutations of NPHS2 may be related with FRNS or SDNS [9,18]. Here, we present for the first time a mutation analysis of NPHS2 and WT1 in children patients lived in western region of KSA, with SRNS, SDNS and FRNS. Table 4 shows a comparison of our findings with those of other studies related to SRNS cases. The mean age of our patients at the onset of NS was 4.5 years, which is similar to that report by Guaragna et al. [30] and Cho et al. [31]. While it was 2.2 years lower in comparison with the African American children patients with SRNS [32]. We found that the male gender may be had a possibility as a risk reason for SRNS, with about 60% in SRNS group. Similar results were reports in a study by Guaragna et al. [30], Chernin et al. [32] and Dhandapani et al. [33].
| This study | Al-Hamed et al. [9] | Kari et al. [34] | Dhandapani et al. [33] | Karle et al. [35] | Guaragna et al. [30] | Cho et al. [31] | |
|---|---|---|---|---|---|---|---|
| Country | KSA | KSA | KSA | India | Germany | Brazil | Korea |
| No. patients | 20 | 62 | 36 | 100 | 31 | 27 | 70 |
| M % | 60 | ND | ND | 62 | 58 | 55.5 | 44.2 |
| F % | 40 | ND | ND | 38 | 42 | 44.5 | 55.8 |
| MN (years) | 4.5 | ND | ND | 5.39 | 3.0 | 4.0 | 4.7 |
| NPHS2 | 15% | 22% | 6.8% | 18% | 28% | 15% | 0% |
| WT1 | 5% | 0% | 0% | ND | ND | 0% | 5.7% |
M: Male; F: Female; MN: Mean age at onset; ND: Not Determined
Table 4. Comparison of NPHS2 and WT1 mutations patterns in the 20 patients of the SRNS in this study with other studies.
Our results revealed that 15% of children with primary SRNS had mutations in the NPHS2. Kari et al. [34] documented the rate of NPHS2 gene mutation in KSA by 6.8%. Several groups from different countries reported that 15%-30% of patients with primary SRNS were caused by mutations in NPHS2 gene [9,30,33,35]. Our results are lower than these frequencies. This could be interpreted by the small number of our patients or possibly reflects ethnic differences reducing the rate of these mutations in Saudi Arabia. It is a well-known that NPHS2 gene mutations are more dominant in Europe and North or South America as the rate of NPHS2 mutations reached to 28% [30,35]. While, Maruyama et al. [36] and Cho et al. [31] did not detect any mutation in NPHS2 within 36 Japanese children or 70 Korean children with SRNS, respectively. Therefore, the variety in the NPHS2 gene mutation frequency is believed to be due to variations in ethnicity and environment [31,36,37].
The nonsense mutation p.Q39X in NPHS2 is the result of c.115C>T transition in exon 1. This mutation had previously been reported [9]. The loss of this amino acid produces a short form of podocin protein. It is supposed that this truncated protein gets stuck in the endoplasmic reticulum, so it loses its ability to bind nephrin in the lipid raft [9,38].
The second heterozygous mutation of NPHS2 gene (c.812C>T; p.P271L) has not been reported in the literature until now. This mutation is located in exon 7 which is considered as a one of c-terminal domain for the NPHS2 protein. It is well-known that the disease-causing mutations of NPHS2 located in the C-terminal domains, which constitute the highly conserved amino acid region of homology with other stomatin protein family, are essential to podocin homo-oligomerization and interaction with other slit diaphragm proteins, especially NPHS1 [5].
In the present study, one gene mutation was found in exon 8 of WT1. This mutation was found in female patient at age 3 months diagnosed as CNS case with renal biopsy of DMS (Table 2). It is a distinguished that NPHS1 is the essential gene that has been recognized in patients presenting CNS in the first 3 months of life [9,13,25]. However, it has also been reported that CNS may be caused by mutations in several other genes, including NPHS2, PLCE1, and WT1 [3,9,16,39]. The low rate of mutation frequency in WT1 gene of our study (5%) are similar to some reports those stated the WT1 mutations may account for about 5% of patients with SRNS, and they have been recognized in patients with isolated DMS, with a clinical onset varying from a few days of life up to 2 years of age [3,16,31]. Interestingly, almost all cases were those of phenotypically female patients, and the mutations occurred mainly in exons 8 and 9, which code for zinc finger domains 2 and 3, respectively [16,31].
We believe that this is the first mutation found in WT1 gene within Saudi children patients with SRNS. Recently, Al-Hamed, et al. [9] performed molecular genetic analysis in a cohort of 49 Saudi Arabian families that included families with CNS, infantile NS and childhood SRNS. They detected mutations in NPHS2, NPHS2, PLCE1 and MYO1E genes, while no mutations were found in WT1 gene [9]. Moreover, Kari et al. [34], retrospectively reviewed 36 children with a clinical diagnosis of SRNS and their results documented that the probably diseasecausing mutations were identified in 5 children, 3 (6.8%) with NPHS2 mutation and 2 (4.5%) had NPHS1 mutation, while there is no mutation found in WT1 gene [34]. Interestingly, the same position of WT1 mutation (c.1301G>A) that found in our patient was considered as a pathogenic mutation in different study that causing the Congenital Diaphragmatic Hernia (CDH), a disorder of the development of the lung and diaphragm that related with pulmonary hypoplasia and pulmonary hypertension [40]. These results confirm the issues about WT1 function as a network complex gene that regulates many functions in human.
Here, NPHS2 or WT1 mutations were not found with SDNS or FRNS patients. Our finding is different from that of Caridi et al. who stated significant heterozygous mutations in children with FRNS and SDNS [18]. Conversely, we detected 9 polymorphic variants, 7 in NPHS2 and 2 in WT1, within the SDNS, FRNS, and SRNS patients. The two silent exonic mutation of exon 1 (c.102A>G) and exon 8 (c.954C>T) in NPHS2 gene were published previously [4].
Interestingly, the SNPs [c.‐51G>T] and [c.-52C>G(;) c.- 51G>T] were identified in the promoter region of NPHS2 gene. The c.‐51G>T variant was published previously, while the (c.-52C>G(;) c.-51G>T) was novel [4]. The Consite web-program predicted the sequences from -55 to -49 (TCCCGTG) as the binding site for the upstream stimulatory factor (USF) in Homo sapiens. Di Duca et al. [41] studied the effect of the regulatory elements in the NPHS2 promoter on the NPHS2 protein function. They reported that the c.‐51G>T variant as a functional polymorphism are affecting the gene expression by 80% down-regulation of the podocin protein when transfected in podocytes [41]. Therefore, the change of nucleotide from G to T in USF binding site resulted in the loss of function of the podocin protein [41]. These facts support the idea that our SRNS patient with the two variants [c.-52C>G (;) c.-51G>T] and SDNS or FRNS patients with the c.‐51G>T variant might be affected through the reduction of the NPHS2 protein level. These facts are in agreement with the published data that suggested children with heterozygous mutations in NPHS2 gene may exhibit the FRNS/SDNS course [18].
We know that the high rate of family history with NS make it difficult to estimate pathogenicity of novel mutations without segregation and extensive family analysis. However, in silico tools have been used to identify pathogenicity of our novel mutations. Similar studies have been used same in silico tools to predict the effect of missense variants with SRNS patients [9,14,19,20,42,43].
Conclusion
We identify 15% of NPHS2 mutations and 5% of WT1 mutation in Saudi SRNS children patients. Interestingly, we detect for the first time a mutation in WT1 gene amongst Saudi children populations. Also, we reported for the first time a haplotype or compound heterozygous mutations [c.-52C>G(;) c.-51G>T] in the promoter region of NPHS2 gene. The low mutations frequencies in the two genes suggest that gene mutations of other podocytespecific proteins could be responsible for the development of SRNS in Saudi children.
Source of Funding
This work was supported by Taif University Fund [grant number 1-435-3645].
Acknowledgement
We gratefully thank all the children and their families at the participating study sites who were enrolled in this study. We thank the technical staff, the nursing, medical assistant and medical staff at the participating hospitals for their help with recruitment of the patients for the study.
References
- Frishberg Y. The genetic basis of steroid-resistant nephrotic syndrome. Paediatr Croat 2015; 59: 44-49.
- Eddy AA, Symons JM. Nephrotic syndrome in childhood. Lancet 2003; 362: 629-639.
- Ruf RG, Schultheiss M, Lichtenberger A, et al. Prevalence of WT1 mutations in a large cohort of patients with steroid-resistant and steroid-sensitive nephrotic syndrome. Kidney Int 2004; 66: 564-570.
- Rachmadi D, Melani A, Monnens L. NPHS2 gene mutation and polymorphisms in Indonesian children with steroid-resistant nephrotic syndrome. J Pediatr 2015; 5: 27-33.
- Sadowski CE, Lovric S, Ashraf S, et al. A single gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol 2015; 26: 1279-1289.
- Kaplan JM, Kim SH, North KN, et al. Mutations in ACTN4, encoding a-actinin-4, cause familial focal segmental glomerulosclerosis. Nat Genet 2000; 24: 251-256.
- Winn MP, Conlon PJ, Lynn KL, et al. A mutation in the TRPC6 cation channel causes familial focal segmental glomerulosclerosis. Science 2005; 308: 1801-1804.
- Brown EJ, Schlöndorff JS, Becker DJ, et al. Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 2010; 42: 2-6.
- Al-Hamed MH, Al-Sabban E, Al-Mojalli H, et al. A molecular genetic analysis of childhood nephrotic syndrome in a cohort of Saudi Arabian families. J Hum Genet 2013; 58: 480-489.
- Gupta IR, Baldwin C, Auguste D, et al. ARHGDIA: A novel gene implicated in nephrotic syndrome. J Med Genet 2013; 50: 330-338.
- Ovunc B, Otto EA, Vega-Warner V, et al. Exome sequencing reveals cubilin mutation as a single gene cause of proteinuria. J Am Soc Nephrol 2011; 22: 1815-1820.
- Sanna-Cherchi S, Burgess KE, Nees SN, et al. Exome sequencing identified MYO1E and NEIL1 as candidate genes for human autosomal recessive steroid-resistant nephrotic syndrome. Kidney Int 2011; 80: 389-396.
- Sako M, Nakanishi K, Obana M, et al. Analysis of NPHS1, NPHS2, ACTN4 and WT1 in Japanese patients with congenital nephrotic syndrome. Kidney Int 2005; 67: 1248-1255.
- SantÃn S, Tazón-Vega B, Silva I, et al. Clinical value of NPHS2 analysis in early- and adult-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Neph 2011; 6: 344-354.
- Machuca E, Hummel A, Nevo F, et al. Clinical and epidemiological assessment of steroid-resistant nephrotic syndrome associated with the NPHS2 R229Q variant. Kidney Int2009; 75: 727-735.
- Mucha B, Ozaltin F, Hinkes BG, et al.Mutations in the Wilms? tumor 1 gene cause isolated steroid resistant nephrotic syndrome and occur in exons 8 and 9. Pediatr Res 2006; 59: 325-331.
- Gbadegesin R, Hinkes B, Vlangos C, et al. Mutational analysis of NPHS2 and WT1 in frequently relapsing and steroid-dependent nephrotic syndrome. Pediatr Nephrol 2007; 22: 509-513.
- Caridi G, Bertelli R, Di Duca M, et al. Broadening the spectrum of diseases related to podocin mutations. J Am Soc Nephrol 2003; 14: 1278-1286.
- Leong IUS, Stuckey A, Lai D, et al. Assessment of the predictive accuracy of five in silico prediction tools, alone or in combination and two metaservers to classify long QT syndrome gene mutations. BMC Med Genet 2015; 16: 34.
- Rodrigues C, Santos-Silva A, Costa E, et al. Performance of in silico tools for the evaluation of UGT1A1 missense variants. Hum Mutat 2015; 36: 1215-1225.
- El Mouzan MI, Al Salloum AA, Al Herbish AS, et al. Consanguinity and major genetic disorders in Saudi children: A community-based cross-sectional study. Ann Saudi Med 2008; 28: 169-173.
- Kari JA. Pediatric renal diseases in the Kingdom of Saudi Arabia. World J Pediatr 2012; 8: 217-221.
- Sharples PM, Poulton J, White RH. Steroid responsive nephrotic syndrome is more common in Asians. Arch Dis Child 1985; 60: 1014-1017.
- Abdurrahman MB, Shipkey FH, Elidrissy AT, et al. Nephrotic syndrome in Saudi infants in the first year of life. Ann Trop Paediatr 1989; 9: 140-146.
- Phadke K, Goodyer P, Bitzan M. Manual of pediatric nephrology. Springer: Berlin, 2014: 141-229.
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248-249.
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012; 7: e46688.
- Wagih OR. Web logo: Plotting custom sequence logos. R package version 1.0.3. 2014.
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 2001; 11: 863-874.
- Guaragna MS, Lutaif AC, Piveta CSC, et al. NPHS2 mutations account for only 15% of nephrotic syndrome cases. BMC Med Genet 2015; 16: 88-96.
- Cho HY, Lee JH, Choi HJ, et al. WT1 and NPHS2 mutations in Korean children with steroid-resistant nephrotic syndrome. Pediatr Nephrol 2008; 23: 63-70.
- Chernin G, Heeringa SF, Gbadegesin R, et al. Low prevalence of NPHS2 mutations in African American children with steroid-resistant nephrotic syndrome. Pediatr Nephrol 2008; 23: 1455-1460.
- Dhandapani MC, Venkatesan V, Rengaswamy NB, et al. Report of novel genetic variation in NPHS2 gene associated with idiopathic nephrotic syndrome in South Indian children. Clin Exp Nephrol 2016.
- Kari JA, El-Desoky SM, Gari M, et al. Steroid-resistant nephrotic syndrome: Impact of genetic testing. Ann Saudi Med 2013; 33: 533-538.
- Karle S, Uetz B, Ronner V, et al. Novel mutations in NPHS2 detected in both familial and sporadic steroid-resistant nephrotic syndrome. J Am Soc Nephrol 2002; 13: 388-393.
- Maruyama K, Iijima K, Ikeda M, et al. NPHS2 mutations in sporadic steroid-resistant nephrotic syndrome in Japanese children. Pediatr Nephrol 2003; 18: 412-416.
- Chanchlani R, Parekh RS. Ethnic differences in childhood nephrotic syndrome. Front Pediatr 2016; 4:39.
- Huber TB, Simons M, Hartleben B, et al. Molecular basis of the functional podocin-nephrin complex: Mutations in the NPHS2 gene disrupt nephrin targeting to lipid raft microdomains. Hum Mol Genet 2003; 12: 3397-3405.
- Hinkes BG, Mucha B, Vlangos CN, et al. Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1 and LAMB2). Pediatrics 2007; 119: e907-e919.
- Antonius T, van Bon B, Eggink A, et al. Denys?Drash syndrome and congenital diaphragmatic hernia: Another case with the 1097G>A (Arg366His) mutation. Am J Med Genet 2008; 146A: 496-499.
- Di Duca M, Oleggini R, Sanna-Cherchi S, et al. Cis and trans regulatory elements in NPHS2 promoter: Implications in proteinuria and progression of renal diseases. Kidney Int 2006; 70: 1332-1341.
- Laurin LP, Lu M, Mottl AK, et al. Podocyte-associated gene mutation screening in a heterogeneous cohort of patients with sporadic focal segmental glomerulosclerosis. Nephrol Dial Transplant 2014; 29: 2062-2069.
- Lipska BS, Balasz-Chmielewska I, Morzuch L, et al. Mutational analysis in podocin-associated hereditary nephrotic syndrome in Polish patients: Founder effect in the Kashubian population. J Appl Genet 2013; 54: 327-333.