Research Article - Biomedical Research (2017) Volume 28, Issue 4
Molecular Identification of Lactobacillus acidophilus as a probiotic potential from traditional doogh samples and evaluation of their antimicrobial activity against some pathogenic bacteria
Vahid Jabbari1,2, Reza Rezaei Mokarram1, Mahmoud Sowti Khiabani1, Fereshteh Askari1, Elham Ahmadi1, Azad mohammad Hassanzadeh1,2, Sanaz buick Aghazadeh2, Mohammad Asgharzadeh3 and Hossein Samadi Kafil2*1Department of Food Science and Technology, Faculty of Agriculture, Tabriz University, Tabriz, Iran
2Drug Applied Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
3Immunology Research Center, Tabriz University of Medical sciences, Tabriz, Iran
- *Corresponding Author:
- Hossein Samadi kafil
Drug Applied Research Center
Tabriz University of Medical Sciences, Iran
Accepted on August 1, 2016
Abstract
Aims: Lactobacillus acidophilus is a potentially probiotic bacteria which added to food products as a probiotic starters to create taste and capabilities such as health-giving and so by this path they enter the food chain of humans. Furthermore, these microorganisms can be identified and isolate in traditional products. In this study tried to identify and isolate the Lactobacillus acidophilus in traditional Doogh samples from Tabriz region in the northwest of Iran.
Material and Methods: The identification of these microorganisms was performed using biochemical tests and molecular method by PCR. Randomly 42 samples were collected from the area, eventually 15 isolates were identified as Lactobacillus acidophilus. To study the antimicrobial activity of cell-free supernatants of isolates against Staphylococcus aureus (ATCC 25923), Shigella dysenteriae and Escherchia coli (ATCC 25922) well diffusion method was used and the inhibitory effect on the growth of pathogenic bacteria was established.
Results and Conclusion: Isolated isolates showed different inhibitory effects so that the highest antimicrobial effect was observed against Shigella dysentery. The results of this study suggest that the isolates isolated from traditional doogh that inhibit the growth of Gram-positive and Gram-negative can be used as new industrial starters.
Keywords
Lactobacillus acidophilus, Probiotic, doogh, Antimicrobial activity.
Introduction
Lactic acid bacteria are Gram-positive microorganisms which are used as starter cultures in fermented dairy products such as yoghurt, cheese, kefir, etc. in food industry [1,2]. Lactobacilluses is an important genus of lactic acid bacteria [3]. More Lactobacillus genus are classified as probiotics which cause health effects on the body. Lactobacillus acidophilus is one of the most common bacterial species with Probiotic activities [4]. This strain contain health-giving properties such as help to lactose digestion, a control factor for blood cholesterol, preventer against cancer and infectious diseases of the digestive system and etc. [5,6]. Researches have shown that Iranian traditional dairies contain bacteria with probiotic properties and with interesting functional properties. Dairy products known as an ideal product for carrying probiotic bacteria to the human intestine [7,8]. The basic problem of probiotic bacteria is that they are fragile under improper environmental conditions which cause loss of their functional properties [9]. Most compounds found in dairy products such as proteins and fats have protective effect on these microorganisms under acidic conditions of the stomach [10]. Doogh is a fermented dairy drink which is highly consumed by Iranian people, a savory yogurt-based beverage popular in Iran and some Middle East countries is one of these products which can contain bacteria with probiotic properties [11]. This dairy product is an Old Iranian acidic dairy drinks that it can be considered as yoghurts drinks with low viscosity. Many studies were done on the isolation and identification of probiotic strains of lactobacillus from different fermented products. In one study, Lactobacillus bacteria of a native product in southern India called idly were identified by PCR (Polymerase Chain Reaction) and antimicrobial properties of these microorganisms was evaluated. The researchers concluded that the isolates from these products can be show suitable fermented property as a starter and also a good bacteriocinic property [12]. In another study, by molecular methods Lactobacillus species were identified with bacteriocin compounds producing feature from clinical samples and food products [13].
The aim of this research was to identification and isolation of Lactobacillus acidophilus from traditional doogh samples, which collected from Tabriz region, and evaluation of the antimicrobial activity of these isolates against some bacteria with pathogenic features. For primary identification, biochemical tests were performed and then molecular method as effective method was used to verify phenotypic identification of Lactobacillus acidophilus from doogh samples.
Material and Methods
Sample collection
40 samples of doogh from the Tabriz area in the Northwest of Iran was collected in icebox and immediately were transported to the laboratory, then kept at 4°C until analysis.
Identification and isolation of Lactobacillus acidophilus
One ml of each sample was added to 9 ml of 0.1% peptone water in test tubes and then dilution steps was conducted by peptone water until 10-5 Dilution. Samples which diluted was cultured on MRS (DE MAN, ROGOSA, and SHARPE) agar medium then incubated at 37°C for 48 h under anaerobic condition [14]. To prevent the growth of yeast in cultures, 0.1% fluconazole was added to cultures. To obtain pure colonies, streak culture was performed on MRS-Sorbitol agar from each colonies which growth on primary MRS medium. After purification of colonies gram staining and biochemical tests were performed [15].
Gram staining test
The colonies grown on culture were examined using gram staining kit according to Collins and colleagues method then observed under light microscope by magnification of 100X [16].
Catalase test
A single isolated colony was used to perform this test. The bacteria picked by loop on a glass slide and one drop of 3% hydrogen peroxide (Merck, Germany) were added on to it. Production of oxygen in reaction indicated the positive response of the bacteria to catalase test. The colonies, which were gram positive and catalase negative, were selected for carbohydrate fermentation test.
Those isolates which appeared to be rod-shaped, Grampositive, non-spore forming, non-pigmented, catalase negative and grown under anaerobic condition were confirmed to belong to the genus lactobacillus.
Carbohydrate fermentation test
The fermentation of carbohydrates (trehalose, raffinose, lactose, galactose, maltose, rhamnose, xylose, mannose, salicin, sucrose, mannitol and cellobiose) was performed according to Bergey's Manual of Systematic Bacteriology to identify all isolated isolates which was gram positive and catalase negative and anaerobically grown at 37°C [17]. For this test Phenol red broth base medium was used.
Molecular identification of Lactobacillus acidophilus
DNA extraction: The DNA of the bacteria was extracted from single colonies which grown on MRS agar (DE MAN, ROGOSA, SHARPE agar, Merck, Darmstadt, Germany) medium [15]. DNA extraction was performed by tissue buffer and according to previous methods [18,19]. Tissue buffer Compounds comprise Sodium Dodecyl Sulfate (SDS) and NaOH. 20 ml of tissue buffer shed in eppendorf and some bacterial colonies add to tissue buffer solution then eppendorf was placed at 95°C on hotplate for 10 min. Finally, solution centrifuged for 1 min at 13000 g and 180 ml of deionized water was added to it, extracted DNAs were taken immediately to -70°C freezer.
PCR reaction and amplification of 16S rRNA gene: 16S rRNA gene fragment specific primers for Lactobacillus acidophilus were purchased from Sinaclon (Sinaclon co, Tehran, Iran). Lactobacillus acidophilus nucleotide sequence specific primers are shown in the Table 1.
| Primer name | nucleotide sequence | Product | Target sequence | Annealing temp. (°C) | References |
|---|---|---|---|---|---|
| Fw | TCTAAGGAAGCGAAGGAT | 273 bp | 16S-23S rRNA | 62 | [21] |
| Rev | CTCTTCTCGGTCGCTCTA | 273 bp | 16S-23S rRNA | 62 |
Table 1. Lactobacillus acidophilus allele- specific primers, which were used in this study.
Reaction mixture consist of Primers, MgCl2, 10X PCR buffer, dNTPs, extracted DNA, distilled water and Taq polymerase in a final volume 25 microliter. The PCR program started with an initial denaturation at 94°C for 4 min, followed by 35 cycles of 94°C for 30 s, 56°C for 45 s and extention step 72°C for 1 min and final extention step was adjusted at 72°C for 4 min [20,21]. PCR products were separated with agarose gel (1.5% w/v) and Amplified fragments visualized by staining with ethidium bromide.
Evaluation of the antimicrobial activity of isolated Lactobacillus acidophilus isolates
In this study, antimicrobial activity of isolated isolates was investigated against pathogenic bacteria such as Staphylococcus aureus ATCC 25923, Shigella dysenteriae and Escherchia coli ATCC 25922. To assess the antibacterial activity of the Lactobacillus acidophilus supernatant, well diffusion assay was performed on Mueller-Hinton agar (Merck, Darmstadt, Germany) [22].
Lactobacillus acidophilus isolates isolated from traditional doogh samples were cultured on MRS-Agar then incubated in anaerobic condition at 37°C for 48 hours. Single colonies were transferred in MRS Broth and incubated for 3 days at 37°C [23]. The culture was then centrifuged at 6,000 xg at 4°C for 20 minutes. Finally, the supernatant was sterilized using 0.2 μm micro-filters to remove any residual bacterial cell. This cell free supernatant was used to evaluate antimicrobial activity.
Furthermore, the pathogenic bacteria were cultured in Tryptose Soy Broth and incubated at 37°C for 24 hours to prepare a turbidity equivalent to 0.5 McFarland standards, approximately to reach Microbial populations about 106 CFU/ml. Then, each pathogen was streaked on the surface of Muller-hinton agar. A sterile Pasture pipette was used to make well (5 mm diameter) on the surface of streaked agar.
Finally, about 100 μl of the prepared supernatant was put inside each well. Plates were incubated at 37°C for 24 hours, after this period, growth inhibitory zone was measured. To increase the sensitivity and accuracy, each test was performed in triplicate and the mean diameter for inhibitory zone was recorded [24].
Statistical analysis
Statistical analyses were performed by descriptive methods and χ2 test (chi-square test). P values below 0.05 were considered statistically significant.
Results
Results obtained by morphological and biochemical tests
From 42 samples, which collected from Tabriz region and cultured on MRS-Sorbitol-Agar, 70 isolate, which was gram positive, were isolated. Tried to select and isolate colonies that were more similar to Lactobacillus acidophilus morphologically under microscope. Between 70 isolates of Gram-positive and rod-shaped colonies, 65 isolates were catalase negative which can degrade catalase and produce bubbles. These isolates was known as Lactobacillus bacteria, stored at -70°C with 15% glycerol, and selected for sugar fermentation test. According to sugar fermentation test, 46% of isolates were identified as Lactobacillus bulgaricus, Lactobacillus acidophilus 25%, Lactobacillus casei 6%, Lactobacillus fermentum 6%, and Lactobacillus plantarum 17%.
Results of Molecular identification of Lactobacillus acidophilus
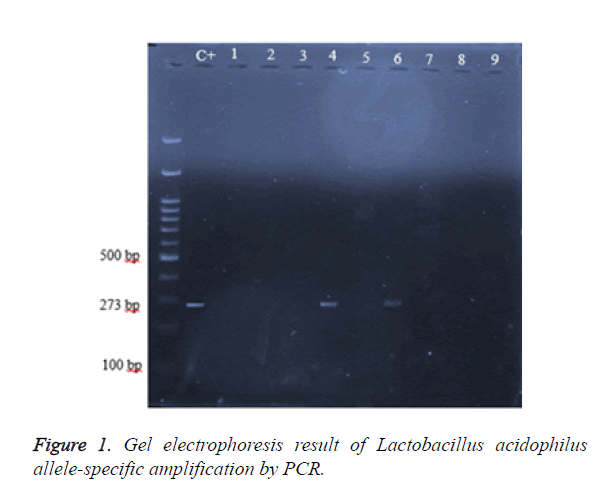
By the results of gel electrophoresis finally 15 isolate identified as Lactobacillus acidophilus. Amplified fragment length was 273 bp (base pair), which is observed in Figure 1. The first well was 100 bp marker or Ladder and next well was Lactobacillus acidophilus positive control, which showed as number 1 and subsequent wells, is detected Lactobacillus acidophiluses on 2, 4 and 6 wells and negative samples in following photos. Considering the high accuracy in molecular identification of Lactobacillus acidophilus the isolates isolated by this method was selected to evaluate the antimicrobial activity.
Antimicrobial effect of Lactobacillus acidophilus isolates isolated from traditional doogh samples against some pathogenic bacteria
To evaluate the antimicrobial activity of isolated isolates against Staphylococcus aureus (ATCC 25923), Shigella dysenteriae and Escherchia coli (ATCC 25922), well diffusion method was used. The inhibition zone was measured and then the results were reported as the mean diameter of inhibition halos in Table 2. Lactobacillus acidophilus, which isolated from traditional doogh samples, had a growth inhibitory activity against pathogenic bacteria, showing different inhibition zone against each one. Thus, the larger halos were observed against Shigella dysenteriae with the mean diameter of inhibition zone about 21.00 mm.
| Isolates/Pathogens | Staphylococcus aureus (mm) | Shigella Dysenteriae (mm) | Escherchia coli (mm) |
|---|---|---|---|
| Lactobacillus acidophilus 1 | 11.6 | 19.14 | 14 |
| Lactobacillus acidophilus 2 | 11.5 | 19.9 | 14.5 |
| Lactobacillus acidophilus 3 | 12 | 20.25 | 13 |
| Lactobacillus acidophilus 4 | 11.3 | 20.2 | 14.7 |
| Lactobacillus acidophilus 5 | 11.5 | 19.1 | 14.5 |
| Lactobacillus acidophilus 6 | 12 | 19.3 | 14.1 |
| Lactobacillus acidophilus 7 | 11 | 20.9 | 15.1 |
| Lactobacillus acidophilus 8 | 11.8 | 20.65 | 14.5 |
| Lactobacillus acidophilus 9 | 10.9 | 19.9 | 13.9 |
| Lactobacillus acidophilus 10 | 11.38 | 20.3 | 14.15 |
| Lactobacillus acidophilus 11 | 10.8 | 21 | 13 |
| Lactobacillus acidophilus 12 | 11.12 | 19.8 | 13.9 |
| Lactobacillus acidophilus 13 | 12.1 | 19.9 | 14.2 |
| Lactobacillus acidophilus 14 | 11.3 | 20.8 | 13.8 |
| Lactobacillus acidophilus 15 | 10.9 | 19 | 13.3 |
Table 2. Mean diameter of inhibition halos of bacteriocin produced against food pathogens.
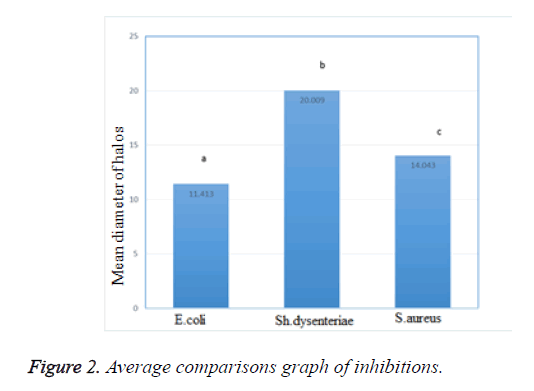
According to average comparisons graph in Figure 2, significant difference between the mean inhibition zone of Shigella dysentery (20/0093) with other bacteria was observed and so significant difference between the mean inhibition zone of Staphylococcus aureus (14/043) with Escherchia coli (11/413) existed (p>0.01).
The results of this study indicate that some Lactobacillus acidophiluses isolated from traditional doogh have significant inhibitory effect on the growth of pathogenic microorganism as shown in Figure 3.
Discussion
Dairy products are an important source to identify and isolate Lactobacilluses. Concerning the health effects of these microorganisms, researchers are always trying to identify and isolate these microorganisms from different products. Primary purpose of this study was to isolate and identify the isolates of the Lactobacillus genus using Psychological, morphological and biochemical tests. Because of these tests from 42 traditional doogh samples, Finally 65 isolates were identified and isolated as Lactobacillus genus. Then this strains used in sugar fermentation test which finally it was found that 46% of isolates were identified as Lactobacillus bulgaricus, Lactobacillus acidophilus 25%, Lactobacillus casei 6%, Lactobacillus fermentum 6%, and Lactobacillus plantarum 17%. Sugar fermentation test showed that Lactobacillus bulgaricus is most abundant isolate in traditional doogh. Considering that, this isolate is the main starter for preparation of yogurt and so doogh, this result was expected.
The main objective of this study was to identification and isolation of Lactobacillus acidophilus as a probiotics potential source from traditional doogh by molecular PCR analysis. By this authentication method, from 65-isolated Lactobacillus genus, finally 15 isolates were identified as Lactobacillus acidophilus by PCR and amplification of the target gene. But the final and most important goal of this study was to determine the antimicrobial activity of Lactobacillus acidophilus isolates which was detected by PCR. All isolates showed antimicrobial effect against the pathogens used in this test. The antimicrobial activity of isolated Lactobacillus acidophilus isolates from traditional doogh was showed weak effect (less than 12 mm) against Escherchia coli. The antimicrobial activity against Staphylococcus aureus strain was reported medium (less than 15 and more than 12 mm). However, greatest antimicrobial effect of strains was reported against Shigella dysentery with an average diameter of inhabitation zone of 20.1 mm. According to average comparisons graph, significant difference was observed between the mean inhibition zone of Shigella dysentery (20/0093) with other bacteria and so significant difference between the mean inhibition zone of Staphylococcus aureus (14/043) with Escherchia coli (11/413) existed (p>0.01). This antimicrobial effect may be inflicted by production of lactic and acetic acids and consequently reduce the pH, diacetyl, hydrogen peroxide, fatty acids, and most importantly may be applied by production of antimicrobial compounds such as bacteriocins.
Conclusion
The results of this study suggest that the isolates isolated from traditional doogh that inhibit the growth of Gram-positive and Gram-negative can be used as new industrial starters.
Acknowledgment
This study was supported by Tabriz University (Faculty of Agriculture) and Drug Applied Research Center, Tabriz University of Medical Sciences. We thank all staff of Microbiology Laboratory of Pashmineh Biomedical Research Center.
References
- Azadnia P, Khan Nazer A. Identification of lactic acid bacteria isolated from traditional drinking yoghurt in tribes of Fars province. Iranian J Veter Res 2009; 10: 235-240.
- Grosu-Tudor SS, Zamfir M, Meulen R, Vuyst L. Isolation of novel homopolysaccharide-producing lactic acid bacteria from Romanian raw milk and fermented dairy products, Eur F Res Technol 2013; 237: 609-615.
- Sade E, Lassila E, Bjorkroth J. Lactic acid bacteria in dried vegetables and spices. Food Microbiol 2016; 53: 110-114.
- Shah NP. Probiotic bacteria: selective enumeration and survival in dairy foods. J Dairy Sci 2000; 83: 894-907.
- Holzapfel WH, Haberer P, Geisen R, Björkroth J, Schillinger U. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am J Clin Nutr 2001; 73: 365S-373S.
- Prasad J, Gill H, Smart J, Gopal PK. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics, Int Dairy J 1998; 8: 993-1002.
- Xiao J, Zhang Y, Yang Z. Lactic acid bacteria in health and disease. Spr Netherlands Dordrecht 2014: 303-374.
- Tamime AY. Probiotic dairy products. John Wiley Sons 2008.
- Sultana K, Godward G, Reynolds N, Arumugaswamy R, Peiris P, Kailasapathy K. Encapsulation of probiotic bacteria with alginate-starch and evaluation of survival in simulated gastrointestinal conditions and in yoghurt, Int J food microbiol 2000; 62: 47-55.
- Madureira AR, Amorim M, Gomes AM, Pintado ME, Malcata FX. Protective effect of whey cheese matrix on probiotic strains exposed to simulated gastrointestinal conditions. Food Res Int 2011; 44: 465-470.
- Asgharzadeh M, Kafil HS, Ebrahimzadeh ME, Bohlouli A. Mannose-binding lectin gene and promoter polymorphism and susceptibility to renal dysfunction in systemic lupus erythematosus. J Biol Sci 2007; 7: 801-805.
- Agaliya PJ, Jeevaratnam K. Molecular characterization of lactobacilli isolated from fermented idli batter. Braz J Microbiol 2014; 44: 1199-1206.
- Ahmad V. Molecular characterization of Bacteriocin Producing Lactobacilli isolated from clinical samples and food products 2016.
- Hugas M, Garriga M, Aymerich T, Monfort JM. Biochemical characterization of lactobacilli from dry fermented sausages. Int J Food Microbiol 1993; 18: 107-113.
- Amin M, Jorfi M, Khosravi A, Samarbafzadeh A, Sheikh AF. Isolation and identification of Lactobacillus casei and Lactobacillus plantarum from plants by PCR and detection of their antibacterial activity, J Biol Sci 2009; 9: 810-814.
- Pyar H, Peh K. Characterization and identification of Lactobacillus acidophilus using biolog rapid identification system. Int J Pharm Pharm Sci 2014; 6: 189-193.
- Holzapfel W, Wood BJ. The genera of lactic acid bacteria, Spr Sci Bus Med 2012.
- Araujo WL, Angellis DAD, Azevedo JL. Direct RAPD evaluation of bacteria without conventional DNA extraction. Brazilian Arch Biol Technol 2004; 47: 375-380.
- Kafil HS, Mobarez AM. Assessment of biofilm formation by enterococci isolates from urinary tract infections with different virulence profiles. J King Saud Univ Sci 2015; 27: 312-317.
- Torriani S, Felis GE, Dellaglio F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers, Applied and Environmental Microbiology 2001; 67: 3450-3454.
- Markiewicz L, Biedrzycka E, Wasilewska E, Bielecka M. Rapid molecular identification and characteristics of Lactobacillus strains. Folia microbiologica 2010; 55: 481-488.
- Moghaddam MZ, Nahaei MR, Kafil HS, Safaeyan F. Bacteriocins of four Lactobacilli species isolated from Yogurt can inhibit growth and Verotoxins production in Escgrecia coli. J Pure Appl Microbiol 2014; 8: 4517-4524.
- Bian L, Molan AL, Maddox I, Shu Q. Antimicrobial activity of Lactobacillus reuteri DPC16 supernatants against selected food borne pathogens. W J Microbiol Biotechnol 2011; 27: 991-998.
- Smaoui S, Elleuch L, Bejar W, Karray-Rebai I, Ayadi I, Jaouadi B, Mathieu F, Chouayekh H, Bejar S, Mellouli L. Inhibition of fungi and gram-negative bacteria by bacteriocin BacTN635 produced by Lactobacillus plantarum sp. TN635, Applied biochemistry and biotechnol 2010; 162: 1132-1146.